Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
APPENDIX I<br />
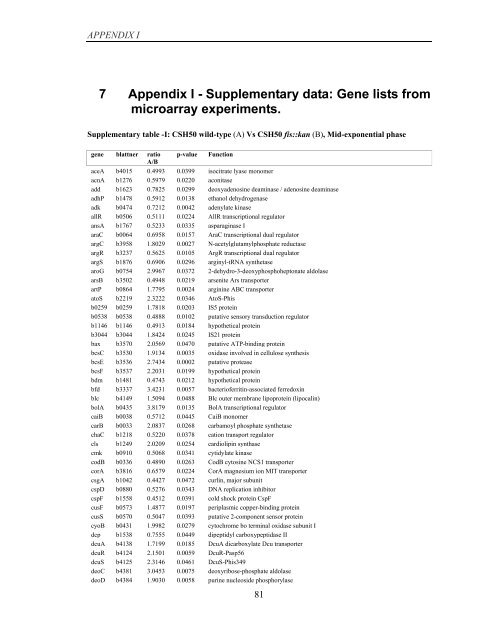
7 Appendix I - Supplementary data: Gene lists from<br />
microarray experiments.<br />
Supplementary table -I: CSH50 wild-type (A) Vs CSH50 fis::kan (B), Mid-exponential phase<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
aceA b4015 0.4993 0.0399 isocitrate lyase monomer<br />
acnA b1276 0.5979 0.0220 aconitase<br />
add b1623 0.7825 0.0299 deoxyadenosine deaminase / adenosine deaminase<br />
adhP b1478 0.5912 0.0138 ethanol dehydrogenase<br />
adk b0474 0.7212 0.0042 adenylate kinase<br />
allR b0506 0.5111 0.0224 AllR transcriptional regulator<br />
ansA b1767 0.5233 0.0335 asparaginase I<br />
araC b0064 0.6958 0.0157 AraC transcriptional dual regulator<br />
argC b3958 1.8029 0.0027 N-acetylglutamylphosphate reductase<br />
argR b3237 0.5625 0.0105 ArgR transcriptional dual regulator<br />
argS b1876 0.6906 0.0296 arginyl-tRNA synthetase<br />
aroG b0754 2.9967 0.0372 2-dehydro-3-deoxyphosphoheptonate aldolase<br />
arsB b3502 0.4948 0.0219 arsenite Ars transporter<br />
artP b0864 1.7795 0.0024 arginine ABC transporter<br />
atoS b2219 2.3222 0.0346 AtoS-Phis<br />
b0259 b0259 1.7818 0.0203 IS5 protein<br />
b0538 b0538 0.4888 0.0102 putative sensory transduction regulator<br />
b1146 b1146 0.4913 0.0184 hypothetical protein<br />
b3044 b3044 1.8424 0.0245 IS21 protein<br />
bax b3570 2.0569 0.0470 putative ATP-binding protein<br />
bcsC b3530 1.9134 0.0035 oxidase involved in cellulose synthesis<br />
bcsE b3536 2.7434 0.0002 putative protease<br />
bcsF b3537 2.2031 0.0199 hypothetical protein<br />
bdm b1481 0.4743 0.0212 hypothetical protein<br />
bfd b3337 3.4231 0.0057 bacteri<strong>of</strong>erritin-associated ferredoxin<br />
blc b4149 1.5094 0.0488 Blc outer membrane lipoprotein (lipocalin)<br />
bolA b0435 3.8179 0.0135 BolA transcriptional regulator<br />
caiB b0038 0.5712 0.0445 CaiB monomer<br />
carB b0033 2.0837 0.0268 carbamoyl phosphate synthetase<br />
chaC b1218 0.5220 0.0378 cation transport regulator<br />
cls b1249 2.0209 0.0254 cardiolipin synthase<br />
cmk b0910 0.5068 0.0341 cytidylate kinase<br />
codB b0336 0.4890 0.0263 CodB cytosine NCS1 transporter<br />
corA b3816 0.6579 0.0224 CorA magnesium ion MIT transporter<br />
csgA b1042 0.4427 0.0472 curlin, major subunit<br />
cspD b0880 0.5276 0.0343 DNA replication inhibitor<br />
cspF b1558 0.4512 0.0391 cold shock protein CspF<br />
cusF b0573 1.4877 0.0197 periplasmic copper-binding protein<br />
cusS b0570 0.5047 0.0393 putative 2-component sensor protein<br />
cyoB b0431 1.9982 0.0279 cytochrome bo terminal oxidase subunit I<br />
dcp b1538 0.7555 0.0449 dipeptidyl carboxypeptidase II<br />
dcuA b4138 1.7199 0.0185 DcuA dicarboxylate Dcu transporter<br />
dcuR b4124 2.1501 0.0059 DcuR-Pasp56<br />
dcuS b4125 2.3146 0.0461 DcuS-Phis349<br />
deoC b4381 3.0453 0.0075 deoxyribose-phosphate aldolase<br />
deoD b4384 1.9030 0.0058 purine nucleoside phosphorylase<br />
81