Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
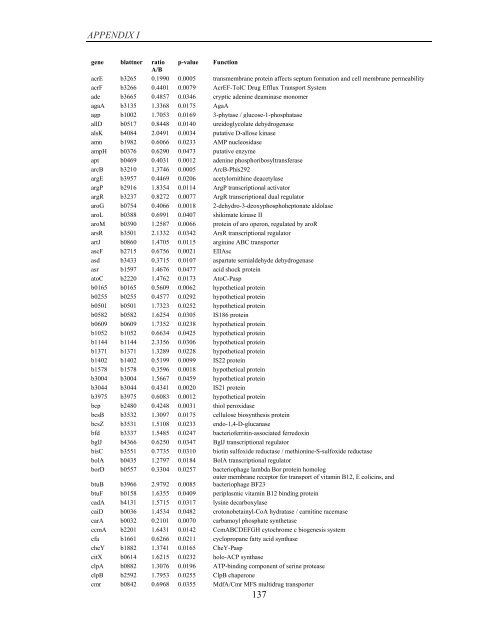
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
acrE b3265 0.1990 0.0005 transmembrane protein affects septum formation and cell membrane permeability<br />
acrF b3266 0.4401 0.0079 AcrEF-TolC Drug Efflux Transport System<br />
ade b3665 0.4857 0.0346 cryptic adenine deaminase monomer<br />
agaA b3135 1.3368 0.0175 AgaA<br />
agp b1002 1.7053 0.0169 3-phytase / glucose-1-phosphatase<br />
allD b0517 0.8448 0.0140 ureidoglycolate dehydrogenase<br />
alsK b4084 2.0491 0.0034 putative D-allose kinase<br />
amn b1982 0.6066 0.0233 AMP nucleosidase<br />
ampH b0376 0.6290 0.0473 putative enzyme<br />
apt b0469 0.4031 0.0012 adenine phosphoribosyltransferase<br />
arcB b3210 1.3746 0.0005 ArcB-Phis292<br />
argE b3957 0.4469 0.0206 acetylornithine deacetylase<br />
argP b2916 1.8354 0.0114 ArgP transcriptional activator<br />
argR b3237 0.8272 0.0077 ArgR transcriptional dual regulator<br />
aroG b0754 0.4066 0.0018 2-dehydro-3-deoxyphosphoheptonate aldolase<br />
aroL b0388 0.6991 0.0407 shikimate kinase II<br />
aroM b0390 1.2587 0.0066 protein <strong>of</strong> aro operon, regulated <strong>by</strong> aroR<br />
arsR b3501 2.1332 0.0342 ArsR transcriptional regulator<br />
artJ b0860 1.4705 0.0115 arginine ABC transporter<br />
ascF b2715 0.6756 0.0021 EIIAsc<br />
asd b3433 0.3715 0.0107 aspartate semialdehyde dehydrogenase<br />
asr b1597 1.4676 0.0477 acid shock protein<br />
atoC b2220 1.4762 0.0173 AtoC-Pasp<br />
b0165 b0165 0.5609 0.0062 hypothetical protein<br />
b0255 b0255 0.4577 0.0292 hypothetical protein<br />
b0501 b0501 1.7323 0.0252 hypothetical protein<br />
b0582 b0582 1.6254 0.0305 IS186 protein<br />
b0609 b0609 1.7352 0.0238 hypothetical protein<br />
b1052 b1052 0.6634 0.0425 hypothetical protein<br />
b1144 b1144 2.3356 0.0306 hypothetical protein<br />
b1371 b1371 1.3289 0.0228 hypothetical protein<br />
b1402 b1402 0.5199 0.0099 IS22 protein<br />
b1578 b1578 0.3596 0.0018 hypothetical protein<br />
b3004 b3004 1.5667 0.0459 hypothetical protein<br />
b3044 b3044 0.4341 0.0020 IS21 protein<br />
b3975 b3975 0.6083 0.0012 hypothetical protein<br />
bcp b2480 0.4248 0.0031 thiol peroxidase<br />
bcsB b3532 1.3097 0.0175 cellulose biosynthesis protein<br />
bcsZ b3531 1.5108 0.0233 endo-1,4-D-glucanase<br />
bfd b3337 1.5485 0.0247 bacteri<strong>of</strong>erritin-associated ferredoxin<br />
bglJ b4366 0.6250 0.0347 BglJ transcriptional regulator<br />
bisC b3551 0.7735 0.0310 biotin sulfoxide reductase / methionine-S-sulfoxide reductase<br />
bolA b0435 1.2797 0.0184 BolA transcriptional regulator<br />
borD b0557 0.3304 0.0257 bacteriophage lambda Bor protein homolog<br />
btuB b3966 2.9792 0.0085<br />
outer membrane receptor for transport <strong>of</strong> vitamin B12, E colicins, and<br />
bacteriophage BF23<br />
btuF b0158 1.6355 0.0409 periplasmic vitamin B12 binding protein<br />
cadA b4131 1.5715 0.0317 lysine decarboxylase<br />
caiD b0036 1.4534 0.0482 crotonobetainyl-CoA hydratase / carnitine racemase<br />
carA b0032 0.2101 0.0070 carbamoyl phosphate synthetase<br />
ccmA b2201 1.6431 0.0142 CcmABCDEFGH cytochrome c bio<strong>gene</strong>sis system<br />
cfa b1661 0.6266 0.0211 cyclopropane fatty acid synthase<br />
cheY b1882 1.3741 0.0165 CheY-Pasp<br />
citX b0614 1.6215 0.0232 holo-ACP synthase<br />
clpA b0882 1.3076 0.0196 ATP-binding component <strong>of</strong> serine protease<br />
clpB b2592 1.7953 0.0255 ClpB chaperone<br />
cmr b0842 0.6968 0.0355 MdfA/Cmr MFS multidrug transporter<br />
137