Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
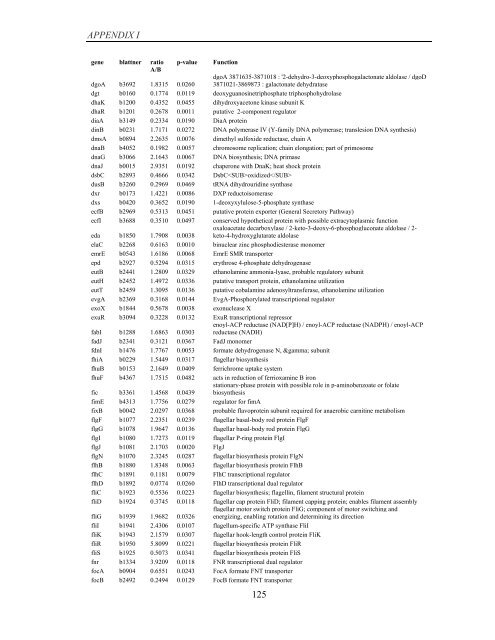
APPENDIX I<br />
<strong>gene</strong> blattner ratio<br />
A/B<br />
p-value<br />
Function<br />
dgoA 3871635-3871018 : '2-dehydro-3-deoxyphosphogalactonate aldolase / dgoD<br />
3871021-3869873 : galactonate dehydratase<br />
dgoA b3692 1.8315 0.0260<br />
dgt b0160 0.1774 0.0119 deoxyguanosinetriphosphate triphosphohydrolase<br />
dhaK b1200 0.4352 0.0455 dihydroxyacetone kinase subunit K<br />
dhaR b1201 0.2678 0.0011 putative 2-component regulator<br />
diaA b3149 0.2334 0.0190 DiaA protein<br />
dinB b0231 1.7171 0.0272 DNA polymerase IV (Y-family DNA polymerase; translesion DNA synthesis)<br />
dmsA b0894 2.2635 0.0076 dimethyl sulfoxide reductase, chain A<br />
dnaB b4052 0.1982 0.0057 chromosome replication; chain elongation; part <strong>of</strong> primosome<br />
dnaG b3066 2.1643 0.0067 DNA biosynthesis; DNA primase<br />
dnaJ b0015 2.9351 0.0192 chaperone with DnaK; heat shock protein<br />
dsbC b2893 0.4666 0.0342 DsbCoxidized<br />
dusB b3260 0.2969 0.0469 tRNA dihydrouridine synthase<br />
dxr b0173 1.4221 0.0086 DXP reductoisomerase<br />
dxs b0420 0.3652 0.0190 1-deoxyxylulose-5-phosphate synthase<br />
ecfB b2969 0.5313 0.0451 putative protein exporter (General Secretory Pathway)<br />
ecfI b3688 0.3510 0.0497 conserved hypothetical protein with possible extracytoplasmic function<br />
oxaloacetate decarboxylase / 2-keto-3-deoxy-6-phosphogluconate aldolase / 2-<br />
eda b1850 1.7908 0.0038 keto-4-hydroxyglutarate aldolase<br />
elaC b2268 0.6163 0.0010 binuclear zinc phosphodiesterase monomer<br />
emrE b0543 1.6186 0.0068 EmrE SMR transporter<br />
epd b2927 0.5294 0.0315 erythrose 4-phosphate dehydrogenase<br />
eutB b2441 1.2809 0.0329 ethanolamine ammonia-lyase, probable regulatory subunit<br />
eutH b2452 1.4972 0.0336 putative transport protein, ethanolamine utilization<br />
eutT b2459 1.3095 0.0136 putative cobalamine adenosyltransferase, ethanolamine utilization<br />
evgA b2369 0.3168 0.0144 EvgA-Phosphorylated transcriptional regulator<br />
exoX b1844 0.5678 0.0038 exonuclease X<br />
exuR b3094 0.3228 0.0132 ExuR transcriptional repressor<br />
enoyl-ACP reductase (NAD[P]H) / enoyl-ACP reductase (NADPH) / enoyl-ACP<br />
fabI b1288 1.6863 0.0303 reductase (NADH)<br />
fadJ b2341 0.3121 0.0367 FadJ monomer<br />
fdnI b1476 1.7767 0.0053 formate dehydrogenase N, γ subunit<br />
fhiA b0229 1.5449 0.0317 flagellar biosynthesis<br />
fhuB b0153 2.1649 0.0409 ferrichrome uptake system<br />
fhuF b4367 1.7515 0.0482 acts in reduction <strong>of</strong> ferrioxamine B iron<br />
stationary-phase protein with possible role in p-aminobenzoate or folate<br />
fic b3361 1.4568 0.0439 biosynthesis<br />
fimE b4313 1.7756 0.0279 regulator for fimA<br />
fixB b0042 2.0297 0.0368 probable flavoprotein subunit required for anaerobic carnitine metabolism<br />
flgF b1077 2.2351 0.0239 flagellar basal-body rod protein FlgF<br />
flgG b1078 1.9647 0.0136 flagellar basal-body rod protein FlgG<br />
flgI b1080 1.7273 0.0119 flagellar P-ring protein FlgI<br />
flgJ b1081 2.1703 0.0020 FlgJ<br />
flgN b1070 2.3245 0.0287 flagellar biosynthesis protein FlgN<br />
flhB b1880 1.8348 0.0063 flagellar biosynthesis protein FlhB<br />
flhC b1891 0.1181 0.0079 FlhC transcriptional regulator<br />
flhD b1892 0.0774 0.0260 FlhD transcriptional dual regulator<br />
fliC b1923 0.5536 0.0223 flagellar biosynthesis; flagellin, filament structural protein<br />
fliD b1924 0.3745 0.0118 flagellar cap protein FliD; filament capping protein; enables filament assembly<br />
flagellar motor switch protein FliG; component <strong>of</strong> motor switching and<br />
fliG b1939 1.9682 0.0326 energizing, enabling rotation and determining its direction<br />
fliI b1941 2.4306 0.0107 flagellum-specific ATP synthase FliI<br />
fliK b1943 2.1579 0.0307 flagellar hook-length control protein FliK<br />
fliR b1950 5.8099 0.0221 flagellar biosynthesis protein FliR<br />
fliS b1925 0.5073 0.0341 flagellar biosynthesis protein FliS<br />
fnr b1334 3.9209 0.0118 FNR transcriptional dual regulator<br />
focA b0904 0.6551 0.0243 FocA formate FNT transporter<br />
focB b2492 0.2494 0.0129 FocB formate FNT transporter<br />
125