Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
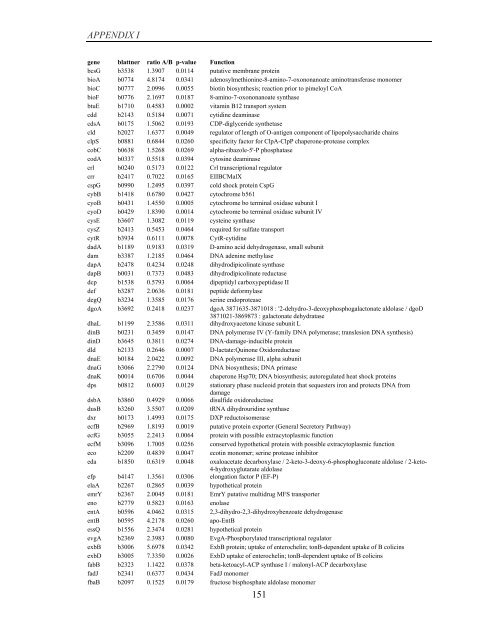
APPENDIX I<br />
<strong>gene</strong> blattner ratio A/B p-value Function<br />
bcsG b3538 1.3907 0.0114 putative membrane protein<br />
bioA b0774 4.8174 0.0341 adenosylmethionine-8-amino-7-oxononanoate aminotransferase monomer<br />
bioC b0777 2.0996 0.0055 biotin biosynthesis; reaction prior to pimeloyl CoA<br />
bioF b0776 2.1697 0.0187 8-amino-7-oxononanoate synthase<br />
btuE b1710 0.4583 0.0002 vitamin B12 transport system<br />
cdd b2143 0.5184 0.0071 cytidine deaminase<br />
cdsA b0175 1.5062 0.0193 CDP-diglyceride synthetase<br />
cld b2027 1.6377 0.0049 regulator <strong>of</strong> length <strong>of</strong> O-antigen component <strong>of</strong> lipopolysaccharide chains<br />
clpS b0881 0.6844 0.0260 specificity factor for ClpA-ClpP chaperone-protease complex<br />
cobC b0638 1.5268 0.0269 alpha-ribazole-5'-P phosphatase<br />
codA b0337 0.5518 0.0394 cytosine deaminase<br />
crl b0240 0.5173 0.0122 Crl transcriptional regulator<br />
crr b2417 0.7022 0.0165 EIIBCMalX<br />
cspG b0990 1.2495 0.0397 cold shock protein CspG<br />
cybB b1418 0.6780 0.0427 cytochrome b561<br />
cyoB b0431 1.4550 0.0005 cytochrome bo terminal oxidase subunit I<br />
cyoD b0429 1.8390 0.0014 cytochrome bo terminal oxidase subunit IV<br />
cysE b3607 1.3082 0.0119 cysteine synthase<br />
cysZ b2413 0.5453 0.0464 required for sulfate transport<br />
cytR b3934 0.6111 0.0078 CytR-cytidine<br />
dadA b1189 0.9183 0.0319 D-amino acid dehydrogenase, small subunit<br />
dam b3387 1.2185 0.0464 DNA adenine methylase<br />
dapA b2478 0.4234 0.0248 dihydrodipicolinate synthase<br />
dapB b0031 0.7373 0.0483 dihydrodipicolinate reductase<br />
dcp b1538 0.5793 0.0064 dipeptidyl carboxypeptidase II<br />
def b3287 2.0636 0.0181 peptide deformylase<br />
degQ b3234 1.3585 0.0176 serine endoprotease<br />
dgoA b3692 0.2418 0.0237 dgoA 3871635-3871018 : '2-dehydro-3-deoxyphosphogalactonate aldolase / dgoD<br />
3871021-3869873 : galactonate dehydratase<br />
dhaL b1199 2.3586 0.0311 dihydroxyacetone kinase subunit L<br />
dinB b0231 0.3459 0.0147 DNA polymerase IV (Y-family DNA polymerase; translesion DNA synthesis)<br />
dinD b3645 0.3811 0.0274 DNA-damage-inducible protein<br />
dld b2133 0.2646 0.0007 D-lactate:Quinone Oxidoreductase<br />
dnaE b0184 2.0422 0.0092 DNA polymerase III, alpha subunit<br />
dnaG b3066 2.2790 0.0124 DNA biosynthesis; DNA primase<br />
dnaK b0014 0.6706 0.0044 chaperone Hsp70; DNA biosynthesis; autoregulated heat shock proteins<br />
dps b0812 0.6003 0.0129 stationary phase nucleoid protein that sequesters iron and protects DNA from<br />
damage<br />
dsbA b3860 0.4929 0.0066 disulfide oxidoreductase<br />
dusB b3260 3.5507 0.0209 tRNA dihydrouridine synthase<br />
dxr b0173 1.4993 0.0175 DXP reductoisomerase<br />
ecfB b2969 1.8193 0.0019 putative protein exporter (General Secretory Pathway)<br />
ecfG b3055 2.2413 0.0064 protein with possible extracytoplasmic function<br />
ecfM b3096 1.7005 0.0256 conserved hypothetical protein with possible extracytoplasmic function<br />
eco b2209 0.4839 0.0047 ecotin monomer; serine protease inhibitor<br />
eda b1850 0.6319 0.0048 oxaloacetate decarboxylase / 2-keto-3-deoxy-6-phosphogluconate aldolase / 2-keto-<br />
4-hydroxyglutarate aldolase<br />
efp b4147 1.3561 0.0306 elongation factor P (EF-P)<br />
elaA b2267 0.2865 0.0039 hypothetical protein<br />
emrY b2367 2.0045 0.0181 EmrY putative multidrug MFS transporter<br />
eno b2779 0.5823 0.0163 enolase<br />
entA b0596 4.0462 0.0315 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase<br />
entB b0595 4.2178 0.0260 apo-EntB<br />
essQ b1556 2.3474 0.0281 hypothetical protein<br />
evgA b2369 2.3983 0.0080 EvgA-Phosphorylated transcriptional regulator<br />
exbB b3006 5.6978 0.0342 ExbB protein; uptake <strong>of</strong> enterochelin; tonB-dependent uptake <strong>of</strong> B colicins<br />
exbD b3005 7.3350 0.0026 ExbD uptake <strong>of</strong> enterochelin; tonB-dependent uptake <strong>of</strong> B colicins<br />
fabB b2323 1.1422 0.0378 beta-ketoacyl-ACP synthase I / malonyl-ACP decarboxylase<br />
fadJ b2341 0.6377 0.0434 FadJ monomer<br />
fbaB b2097 0.1525 0.0179 fructose bisphosphate aldolase monomer<br />
151