Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
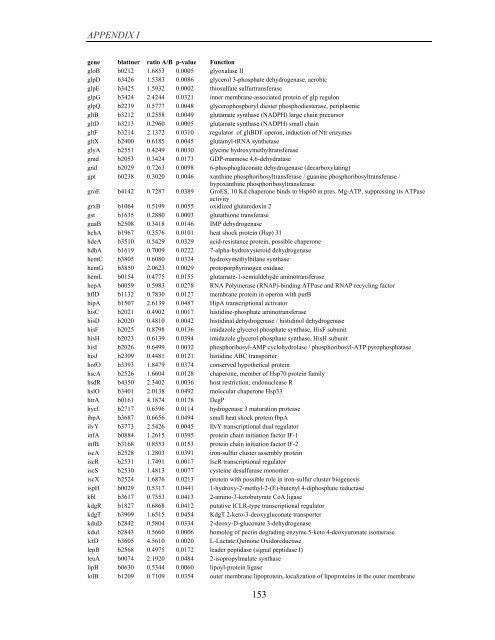
APPENDIX I<br />
<strong>gene</strong> blattner ratio A/B p-value Function<br />
gloB b0212 1.6853 0.0005 glyoxalase II<br />
glpD b3426 1.5383 0.0086 glycerol 3-phosphate dehydrogenase, aerobic<br />
glpE b3425 1.5932 0.0002 thiosulfate sulfurtransferase<br />
glpG b3424 2.4244 0.0321 inner membrane-associated protein <strong>of</strong> glp regulon<br />
glpQ b2239 0.5777 0.0048 glycerophosphoryl diester phosphodiesterase, periplasmic<br />
gltB b3212 0.2558 0.0049 glutamate synthase (NADPH) large chain precursor<br />
gltD b3213 0.2960 0.0005 glutamate synthase (NADPH) small chain<br />
gltF b3214 2.1372 0.0310 regulator <strong>of</strong> gltBDF operon, induction <strong>of</strong> Ntr enzymes<br />
gltX b2400 0.6185 0.0045 glutamyl-tRNA synthetase<br />
glyA b2551 0.4249 0.0030 glycine hydroxymethyltransferase<br />
gmd b2053 0.3424 0.0173 GDP-mannose 4,6-dehydratase<br />
gnd b2029 0.7263 0.0098 6-phosphogluconate dehydrogenase (decarboxylating)<br />
gpt b0238 0.3020 0.0046 xanthine phosphoribosyltransferase / guanine phosphoribosyltransferase /<br />
hypoxanthine phosphoribosyltransferase<br />
groE b4142 0.7287 0.0389 GroES, 10 Kd chaperone binds to Hsp60 in pres. Mg-ATP, suppressing its ATPase<br />
activity<br />
grxB b1064 0.5199 0.0055 oxidized glutaredoxin 2<br />
gst b1635 0.2880 0.0003 glutathione transferase<br />
guaB b2508 0.3418 0.0146 IMP dehydrogenase<br />
hchA b1967 0.3576 0.0101 heat shock protein (Hsp) 31<br />
hdeA b3510 0.5429 0.0329 acid-resistance protein, possible chaperone<br />
hdhA b1619 0.7009 0.0222 7-alpha-hydroxysteroid dehydrogenase<br />
hemC b3805 0.6080 0.0324 hydroxymethylbilane synthase<br />
hemG b3850 2.0623 0.0029 protoporphyrinogen oxidase<br />
hemL b0154 0.4775 0.0155 glutamate-1-semialdehyde aminotransferase<br />
hepA b0059 0.5983 0.0278 RNA Polymerase (RNAP)-binding ATPase and RNAP recycling factor<br />
hflD b1132 0.7830 0.0127 membrane protein in operon with purB<br />
hipA b1507 2.6139 0.0487 HipA transcriptional activator<br />
hisC b2021 0.4902 0.0017 histidine-phosphate aminotransferase<br />
hisD b2020 0.4810 0.0042 histidinal dehydrogenase / histidinol dehydrogenase<br />
hisF b2025 0.8798 0.0136 imidazole glycerol phosphate synthase, HisF subunit<br />
hisH b2023 0.6139 0.0394 imidazole glycerol phosphate synthase, HisH subunit<br />
hisI b2026 0.6499 0.0032 phosphoribosyl-AMP cyclohydrolase / phosphoribosyl-ATP pyrophosphatase<br />
hisJ b2309 0.4481 0.0121 histidine ABC transporter<br />
h<strong>of</strong>O b3393 1.8479 0.0374 conserved hypothetical protein<br />
hscA b2526 1.6604 0.0128 chaperone, member <strong>of</strong> Hsp70 protein family<br />
hsdR b4350 2.3402 0.0036 host restriction; endonuclease R<br />
hslO b3401 2.0138 0.0492 molecular chaperone Hsp33<br />
htrA b0161 4.1874 0.0178 DegP<br />
hycI b2717 0.6596 0.0114 hydrogenase 3 maturation protease<br />
ibpA b3687 0.6656 0.0494 small heat shock protein IbpA<br />
ilvY b3773 2.5426 0.0045 IlvY transcriptional dual regulator<br />
infA b0884 1.2615 0.0395 protein chain initiation factor IF-1<br />
infB b3168 0.8553 0.0153 protein chain initiation factor IF-2<br />
iscA b2528 1.2803 0.0391 iron-sulfur cluster assembly protein<br />
iscR b2531 1.7491 0.0017 IscR transcriptional regulator<br />
iscS b2530 1.4813 0.0077 cysteine desulfurase monomer<br />
iscX b2524 1.6876 0.0213 protein with possible role in iron-sulfur cluster bio<strong>gene</strong>sis<br />
ispH b0029 0.5317 0.0441 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase<br />
kbl b3617 0.7553 0.0413 2-amino-3-ketobutyrate CoA ligase<br />
kdgR b1827 0.6868 0.0412 putative ICLR-type transcriptional regulator<br />
kdgT b3909 1.6515 0.0454 KdgT 2-keto-3-deoxygluconate transporter<br />
kduD b2842 0.5804 0.0334 2-deoxy-D-gluconate 3-dehydrogenase<br />
kduI b2843 0.5660 0.0006 homolog <strong>of</strong> pectin degrading enzyme 5-keto 4-deoxyuronate isomerase<br />
lctD b3605 4.5610 0.0020 L-Lactate:Quinone Oxidoreductase<br />
lepB b2568 0.4975 0.0172 leader peptidase (signal peptidase I)<br />
leuA b0074 2.1920 0.0484 2-isopropylmalate synthase<br />
lipB b0630 0.5344 0.0060 lipoyl-protein ligase<br />
lolB b1209 0.7109 0.0354 outer membrane lipoprotein, localization <strong>of</strong> lipoproteins in the outer membrane<br />
153