Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
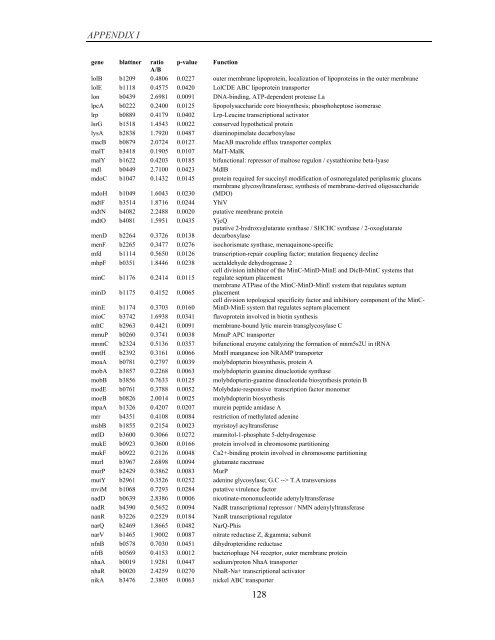
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
lolB b1209 0.4806 0.0227 outer membrane lipoprotein, localization <strong>of</strong> lipoproteins in the outer membrane<br />
lolE b1118 0.4575 0.0420 LolCDE ABC lipoprotein transporter<br />
lon b0439 2.6981 0.0091 DNA-binding, ATP-dependent protease La<br />
lpcA b0222 0.2400 0.0125 lipopolysaccharide core biosynthesis; phosphoheptose isomerase<br />
lrp b0889 0.4179 0.0402 Lrp-Leucine transcriptional activator<br />
lsrG b1518 1.4543 0.0022 conserved hypothetical protein<br />
lysA b2838 1.7920 0.0487 diaminopimelate decarboxylase<br />
macB b0879 2.0724 0.0127 MacAB macrolide efflux transporter complex<br />
malT b3418 0.1905 0.0107 MalT-MalK<br />
malY b1622 0.4203 0.0185 bifunctional: repressor <strong>of</strong> maltose regulon / cystathionine beta-lyase<br />
mdl b0449 2.7100 0.0423 MdlB<br />
mdoC b1047 0.1432 0.0145 protein required for succinyl modification <strong>of</strong> osmoregulated periplasmic glucans<br />
mdoH b1049 1.6043 0.0230<br />
membrane glycosyltransferase; synthesis <strong>of</strong> membrane-derived oligosaccharide<br />
(MDO)<br />
mdtF b3514 1.8716 0.0244 YhiV<br />
mdtN b4082 2.2488 0.0020 putative membrane protein<br />
mdtO b4081 1.5951 0.0435 YjcQ<br />
menD b2264 0.3726 0.0138<br />
putative 2-hydroxyglutarate synthase / SHCHC synthase / 2-oxoglutarate<br />
decarboxylase<br />
menF b2265 0.3477 0.0276 isochorismate synthase, menaquinone-specific<br />
mfd b1114 0.5650 0.0126 transcription-repair coupling factor; mutation frequency decline<br />
mhpF b0351 1.8446 0.0238 acetaldehyde dehydrogenase 2<br />
minC b1176 0.2414 0.0115<br />
cell division inhibitor <strong>of</strong> the MinC-MinD-MinE and DicB-MinC systems that<br />
regulate septum placement<br />
minD b1175 0.4152 0.0065<br />
membrane ATPase <strong>of</strong> the MinC-MinD-MinE system that regulates septum<br />
placement<br />
minE b1174 0.3703 0.0160<br />
cell division topological specificity factor and inhibitory component <strong>of</strong> the MinC-<br />
MinD-MinE system that regulates septum placement<br />
mioC b3742 1.6938 0.0341 flavoprotein involved in biotin synthesis<br />
mltC b2963 0.4421 0.0091 membrane-bound lytic murein transglycosylase C<br />
mmuP b0260 0.3741 0.0038 MmuP APC transporter<br />
mnmC b2324 0.5136 0.0357 bifunctional enzyme catalyzing the formation <strong>of</strong> mnm5s2U in tRNA<br />
mntH b2392 0.3161 0.0066 MntH manganese ion NRAMP transporter<br />
moaA b0781 0.2797 0.0039 molybdopterin biosynthesis, protein A<br />
mobA b3857 0.2268 0.0063 molybdopterin guanine dinucleotide synthase<br />
mobB b3856 0.7633 0.0125 molybdopterin-guanine dinucleotide biosynthesis protein B<br />
modE b0761 0.3788 0.0052 Molybdate-responsive transcription factor monomer<br />
moeB b0826 2.0014 0.0025 molybdopterin biosynthesis<br />
mpaA b1326 0.4207 0.0207 murein peptide amidase A<br />
mrr b4351 0.4108 0.0084 restriction <strong>of</strong> methylated adenine<br />
msbB b1855 0.2154 0.0023 myristoyl acyltransferase<br />
mtlD b3600 0.3066 0.0272 mannitol-1-phosphate 5-dehydrogenase<br />
mukE b0923 0.3600 0.0166 protein involved in chromosome partitioning<br />
mukF b0922 0.2126 0.0048 Ca2+-binding protein involved in chromosome partitioning<br />
murI b3967 2.6898 0.0094 glutamate racemase<br />
murP b2429 0.3862 0.0083 MurP<br />
mutY b2961 0.3526 0.0252 adenine glycosylase; G.C --> T.A transversions<br />
mviM b1068 0.7293 0.0284 putative virulence factor<br />
nadD b0639 2.8386 0.0006 nicotinate-mononucleotide adenylyltransferase<br />
nadR b4390 0.5652 0.0094 NadR transcriptional repressor / NMN adenylyltransferase<br />
nanR b3226 0.2529 0.0184 NanR transcriptional regulator<br />
narQ b2469 1.8665 0.0482 NarQ-Phis<br />
narV b1465 1.9002 0.0087 nitrate reductase Z, γ subunit<br />
nfnB b0578 0.7030 0.0451 dihydropteridine reductase<br />
nfrB b0569 0.4153 0.0012 bacteriophage N4 receptor, outer membrane protein<br />
nhaA b0019 1.9281 0.0447 sodium/proton NhaA transporter<br />
nhaR b0020 2.4259 0.0270 NhaR-Na+ transcriptional activator<br />
nikA b3476 2.3805 0.0063 nickel ABC transporter<br />
128