Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
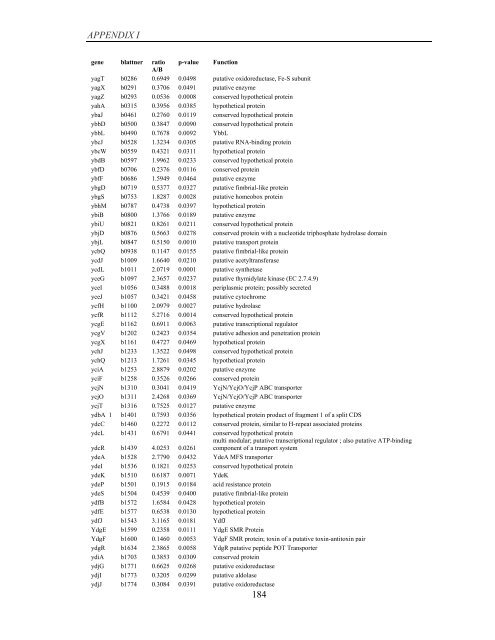
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
yagT b0286 0.6949 0.0498 putative oxidoreductase, Fe-S subunit<br />
yagX b0291 0.3706 0.0491 putative enzyme<br />
yagZ b0293 0.0536 0.0008 conserved hypothetical protein<br />
yahA b0315 0.3956 0.0385 hypothetical protein<br />
ybaJ b0461 0.2760 0.0119 conserved hypothetical protein<br />
ybbD b0500 0.3847 0.0090 conserved hypothetical protein<br />
ybbL b0490 0.7678 0.0092 YbbL<br />
ybcJ b0528 1.3234 0.0305 putative RNA-binding protein<br />
ybcW b0559 0.4321 0.0311 hypothetical protein<br />
ybdB b0597 1.9962 0.0233 conserved hypothetical protein<br />
ybfD b0706 0.2376 0.0116 conserved protein<br />
ybfF b0686 1.5949 0.0464 putative enzyme<br />
ybgD b0719 0.5377 0.0327 putative fimbrial-like protein<br />
ybgS b0753 1.8287 0.0028 putative homeobox protein<br />
ybhM b0787 0.4738 0.0397 hypothetical protein<br />
ybiB b0800 1.3766 0.0189 putative enzyme<br />
ybiU b0821 0.8261 0.0211 conserved hypothetical protein<br />
ybjD b0876 0.5663 0.0278 conserved protein with a nucleotide triphosphate hydrolase domain<br />
ybjL b0847 0.5150 0.0010 putative transport protein<br />
ycbQ b0938 0.1147 0.0155 putative fimbrial-like protein<br />
ycdJ b1009 1.6640 0.0210 putative acetyltransferase<br />
ycdL b1011 2.0719 0.0001 putative synthetase<br />
yceG b1097 2.3657 0.0237 putative thymidylate kinase (EC 2.7.4.9)<br />
yceI b1056 0.3488 0.0018 periplasmic protein; possibly secreted<br />
yceJ b1057 0.3421 0.0458 putative cytochrome<br />
ycfH b1100 2.0979 0.0027 putative hydrolase<br />
ycfR b1112 5.2716 0.0014 conserved hypothetical protein<br />
ycgE b1162 0.6911 0.0063 putative transcriptional regulator<br />
ycgV b1202 0.2423 0.0354 putative adhesion and penetration protein<br />
ycgX b1161 0.4727 0.0469 hypothetical protein<br />
ychJ b1233 1.3522 0.0498 conserved hypothetical protein<br />
ychQ b1213 1.7261 0.0345 hypothetical protein<br />
yciA b1253 2.8879 0.0202 putative enzyme<br />
yciF b1258 0.3526 0.0266 conserved protein<br />
ycjN b1310 0.3041 0.0419 YcjN/YcjO/YcjP ABC transporter<br />
ycjO b1311 2.4268 0.0369 YcjN/YcjO/YcjP ABC transporter<br />
ycjT b1316 0.7525 0.0127 putative enzyme<br />
ydbA_1 b1401 0.7593 0.0356 hypothetical protein product <strong>of</strong> fragment 1 <strong>of</strong> a split CDS<br />
ydcC b1460 0.2272 0.0112 conserved protein, similar to H-repeat associated proteins<br />
ydcL b1431 0.6791 0.0441 conserved hypothetical protein<br />
ydcR b1439 4.0253 0.0261<br />
multi modular; putative transcriptional regulator ; also putative ATP-binding<br />
component <strong>of</strong> a transport system<br />
ydeA b1528 2.7790 0.0432 YdeA MFS transporter<br />
ydeI b1536 0.1821 0.0253 conserved hypothetical protein<br />
ydeK b1510 0.6187 0.0071 YdeK<br />
ydeP b1501 0.1915 0.0184 acid resistance protein<br />
ydeS b1504 0.4539 0.0400 putative fimbrial-like protein<br />
ydfB b1572 1.6584 0.0428 hypothetical protein<br />
ydfE b1577 0.6538 0.0130 hypothetical protein<br />
ydfJ b1543 3.1165 0.0181 YdfJ<br />
YdgE b1599 0.2358 0.0111 YdgE SMR Protein<br />
YdgF b1600 0.1460 0.0053 YdgF SMR protein; toxin <strong>of</strong> a putative toxin-antitoxin pair<br />
ydgR b1634 2.3865 0.0058 YdgR putative peptide POT Transporter<br />
ydiA b1703 0.3853 0.0309 conserved protein<br />
ydjG b1771 0.6625 0.0268 putative oxidoreductase<br />
ydjI b1773 0.3205 0.0299 putative aldolase<br />
ydjJ b1774 0.3084 0.0391 putative oxidoreductase<br />
184