Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
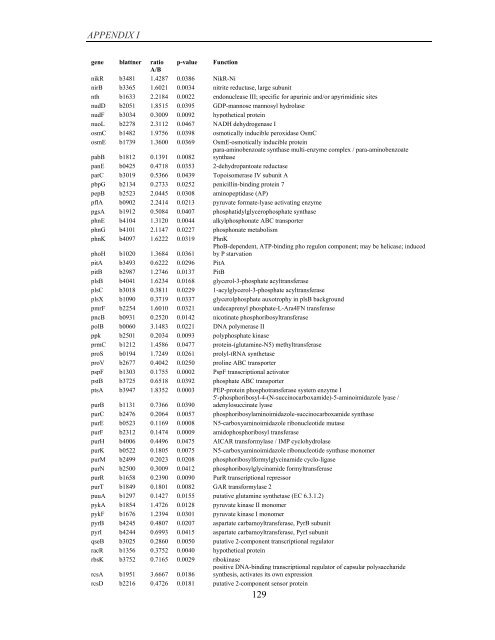
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
nikR b3481 1.4287 0.0386 NikR-Ni<br />
nirB b3365 1.6021 0.0034 nitrite reductase, large subunit<br />
nth b1633 2.2184 0.0022 endonuclease III; specific for apurinic and/or apyrimidinic sites<br />
nudD b2051 1.8515 0.0395 GDP-mannose mannosyl hydrolase<br />
nudF b3034 0.3009 0.0092 hypothetical protein<br />
nuoL b2278 2.3112 0.0467 NADH dehydrogenase I<br />
osmC b1482 1.9756 0.0398 osmotically inducible peroxidase OsmC<br />
osmE b1739 1.3600 0.0369 OsmE-osmotically inducible protein<br />
pabB b1812 0.1391 0.0082<br />
para-aminobenzoate synthase multi-enzyme complex / para-aminobenzoate<br />
synthase<br />
panE b0425 0.4718 0.0353 2-dehydropantoate reductase<br />
parC b3019 0.5366 0.0439 Topoisomerase IV subunit A<br />
pbpG b2134 0.2733 0.0252 penicillin-binding protein 7<br />
pepB b2523 2.0445 0.0308 aminopeptidase (AP)<br />
pflA b0902 2.2414 0.0213 pyruvate formate-lyase activating enzyme<br />
pgsA b1912 0.5084 0.0407 phosphatidylglycerophosphate synthase<br />
phnE b4104 1.3120 0.0044 alkylphosphonate ABC transporter<br />
phnG b4101 2.1147 0.0227 phosphonate metabolism<br />
phnK b4097 1.6222 0.0319 PhnK<br />
phoH b1020 1.3684 0.0361<br />
PhoB-dependent, ATP-binding pho regulon component; may be helicase; induced<br />
<strong>by</strong> P starvation<br />
pitA b3493 0.6222 0.0296 PitA<br />
pitB b2987 1.2746 0.0137 PitB<br />
plsB b4041 1.6234 0.0168 glycerol-3-phosphate acyltransferase<br />
plsC b3018 0.3811 0.0229 1-acylglycerol-3-phosphate acyltransferase<br />
plsX b1090 0.3719 0.0337 glycerolphosphate auxotrophy in plsB background<br />
pmrF b2254 1.6010 0.0321 undecaprenyl phosphate-L-Ara4FN transferase<br />
pncB b0931 0.2520 0.0142 nicotinate phosphoribosyltransferase<br />
polB b0060 3.1483 0.0221 DNA polymerase II<br />
ppk b2501 0.2034 0.0093 polyphosphate kinase<br />
prmC b1212 1.4586 0.0477 protein-(glutamine-N5) methyltransferase<br />
proS b0194 1.7249 0.0261 prolyl-tRNA synthetase<br />
proV b2677 0.4042 0.0250 proline ABC transporter<br />
pspF b1303 0.1755 0.0002 PspF transcriptional activator<br />
pstB b3725 0.6518 0.0392 phosphate ABC transporter<br />
ptsA b3947 1.8352 0.0003 PEP-protein phosphotransferase system enzyme I<br />
purB b1131 0.7366 0.0390<br />
5'-phosphoribosyl-4-(N-succinocarboxamide)-5-aminoimidazole lyase /<br />
adenylosuccinate lyase<br />
purC b2476 0.2064 0.0057 phosphoribosylaminoimidazole-succinocarboxamide synthase<br />
purE b0523 0.1169 0.0008 N5-carboxyaminoimidazole ribonucleotide mutase<br />
purF b2312 0.1474 0.0009 amidophosphoribosyl transferase<br />
purH b4006 0.4496 0.0475 AICAR transformylase / IMP cyclohydrolase<br />
purK b0522 0.1805 0.0075 N5-carboxyaminoimidazole ribonucleotide synthase monomer<br />
purM b2499 0.2023 0.0208 phosphoribosylformylglycinamide cyclo-ligase<br />
purN b2500 0.3009 0.0412 phosphoribosylglycinamide formyltransferase<br />
purR b1658 0.2390 0.0090 PurR transcriptional repressor<br />
purT b1849 0.1801 0.0082 GAR transformylase 2<br />
puuA b1297 0.1427 0.0155 putative glutamine synthetase (EC 6.3.1.2)<br />
pykA b1854 1.4726 0.0128 pyruvate kinase II monomer<br />
pykF b1676 1.2394 0.0301 pyruvate kinase I monomer<br />
pyrB b4245 0.4807 0.0207 aspartate carbamoyltransferase, PyrB subunit<br />
pyrI b4244 0.6993 0.0415 aspartate carbamoyltransferase, PyrI subunit<br />
qseB b3025 0.2860 0.0050 putative 2-component transcriptional regulator<br />
racR b1356 0.3752 0.0040 hypothetical protein<br />
rbsK b3752 0.7165 0.0029 ribokinase<br />
rcsA b1951 3.6667 0.0186<br />
positive DNA-binding transcriptional regulator <strong>of</strong> capsular polysaccharide<br />
synthesis, activates its own <strong>expression</strong><br />
rcsD b2216 0.4726 0.0181 putative 2-component sensor protein<br />
129