Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
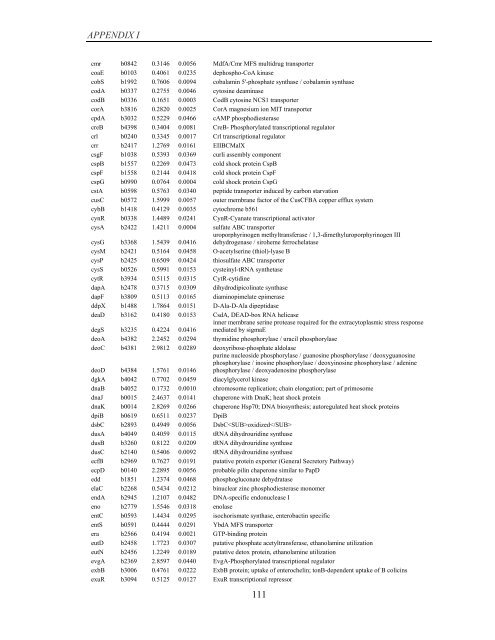
APPENDIX I<br />
cmr b0842 0.3146 0.0056 MdfA/Cmr MFS multidrug transporter<br />
coaE b0103 0.4061 0.0235 dephospho-CoA kinase<br />
cobS b1992 0.7606 0.0094 cobalamin 5'-phosphate synthase / cobalamin synthase<br />
codA b0337 0.2755 0.0046 cytosine deaminase<br />
codB b0336 0.1651 0.0003 CodB cytosine NCS1 transporter<br />
corA b3816 0.2820 0.0025 CorA magnesium ion MIT transporter<br />
cpdA b3032 0.5229 0.0466 cAMP phosphodiesterase<br />
creB b4398 0.3404 0.0081 CreB- Phosphorylated transcriptional regulator<br />
crl b0240 0.3345 0.0017 Crl transcriptional regulator<br />
crr b2417 1.2769 0.0161 EIIBCMalX<br />
csgF b1038 0.5393 0.0369 curli assembly component<br />
cspB b1557 0.2269 0.0473 cold shock protein CspB<br />
cspF b1558 0.2144 0.0418 cold shock protein CspF<br />
cspG b0990 0.0764 0.0004 cold shock protein CspG<br />
cstA b0598 0.5763 0.0340 peptide transporter induced <strong>by</strong> carbon starvation<br />
cusC b0572 1.5999 0.0057 outer membrane factor <strong>of</strong> the CusCFBA copper efflux system<br />
cybB b1418 0.4129 0.0035 cytochrome b561<br />
cynR b0338 1.4489 0.0241 CynR-Cyanate transcriptional activator<br />
cysA b2422 1.4211 0.0004 sulfate ABC transporter<br />
cysG b3368 1.5439 0.0416<br />
uroporphyrinogen methyltransferase / 1,3-dimethyluroporphyrinogen III<br />
dehydrogenase / siroheme ferrochelatase<br />
cysM b2421 0.5164 0.0458 O-acetylserine (thiol)-lyase B<br />
cysP b2425 0.6509 0.0424 thiosulfate ABC transporter<br />
cysS b0526 0.5991 0.0153 cysteinyl-tRNA synthetase<br />
cytR b3934 0.5115 0.0315 CytR-cytidine<br />
dapA b2478 0.3715 0.0309 dihydrodipicolinate synthase<br />
dapF b3809 0.5113 0.0165 diaminopimelate epimerase<br />
ddpX b1488 1.7864 0.0151 D-Ala-D-Ala dipeptidase<br />
deaD b3162 0.4180 0.0153 CsdA, DEAD-box RNA helicase<br />
degS b3235 0.4224 0.0416<br />
inner membrane serine protease required for the extracytoplasmic stress response<br />
mediated <strong>by</strong> sigmaE<br />
deoA b4382 2.2452 0.0294 thymidine phosphorylase / uracil phosphorylase<br />
deoC b4381 2.9812 0.0289 deoxyribose-phosphate aldolase<br />
deoD b4384 1.5761 0.0146<br />
purine nucleoside phosphorylase / guanosine phosphorylase / deoxyguanosine<br />
phosphorylase / inosine phosphorylase / deoxyinosine phosphorylase / adenine<br />
phosphorylase / deoxyadenosine phosphorylase<br />
dgkA b4042 0.7702 0.0459 diacylglycerol kinase<br />
dnaB b4052 0.1732 0.0010 chromosome replication; chain elongation; part <strong>of</strong> primosome<br />
dnaJ b0015 2.4637 0.0141 chaperone with DnaK; heat shock protein<br />
dnaK b0014 2.8269 0.0266 chaperone Hsp70; DNA biosynthesis; autoregulated heat shock proteins<br />
dpiB b0619 0.6511 0.0237 DpiB<br />
dsbC b2893 0.4949 0.0056 DsbCoxidized<br />
dusA b4049 0.4059 0.0115 tRNA dihydrouridine synthase<br />
dusB b3260 0.8122 0.0209 tRNA dihydrouridine synthase<br />
dusC b2140 0.5406 0.0092 tRNA dihydrouridine synthase<br />
ecfB b2969 0.7627 0.0191 putative protein exporter (General Secretory Pathway)<br />
ecpD b0140 2.2895 0.0056 probable pilin chaperone similar to PapD<br />
edd b1851 1.2374 0.0468 phosphogluconate dehydratase<br />
elaC b2268 0.5434 0.0212 binuclear zinc phosphodiesterase monomer<br />
endA b2945 1.2107 0.0482 DNA-specific endonuclease I<br />
eno b2779 1.5546 0.0318 enolase<br />
entC b0593 1.4434 0.0295 isochorismate synthase, enterobactin specific<br />
entS b0591 0.4444 0.0291 YbdA MFS transporter<br />
era b2566 0.4194 0.0021 GTP-binding protein<br />
eutD b2458 1.7723 0.0307 putative phosphate acetyltransferase, ethanolamine utilization<br />
eutN b2456 1.2249 0.0189 putative detox protein, ethanolamine utilization<br />
evgA b2369 2.8597 0.0440 EvgA-Phosphorylated transcriptional regulator<br />
exbB b3006 0.4761 0.0222 ExbB protein; uptake <strong>of</strong> enterochelin; tonB-dependent uptake <strong>of</strong> B colicins<br />
exuR b3094 0.5125 0.0127 ExuR transcriptional repressor<br />
111