Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
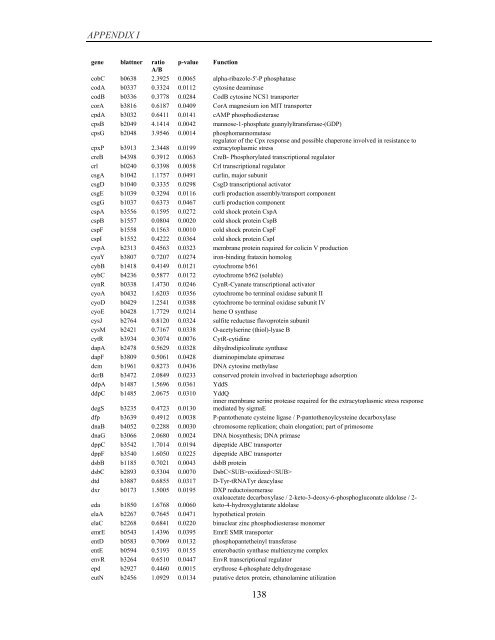
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
cobC b0638 2.3925 0.0065 alpha-ribazole-5'-P phosphatase<br />
codA b0337 0.3324 0.0112 cytosine deaminase<br />
codB b0336 0.3778 0.0284 CodB cytosine NCS1 transporter<br />
corA b3816 0.6187 0.0409 CorA magnesium ion MIT transporter<br />
cpdA b3032 0.6411 0.0141 cAMP phosphodiesterase<br />
cpsB b2049 4.1414 0.0042 mannose-1-phosphate guanylyltransferase-(GDP)<br />
cpsG b2048 3.9546 0.0014 phosphomannomutase<br />
cpxP b3913 2.3448 0.0199<br />
regulator <strong>of</strong> the Cpx response and possible chaperone involved in resistance to<br />
extracytoplasmic stress<br />
creB b4398 0.3912 0.0063 CreB- Phosphorylated transcriptional regulator<br />
crl b0240 0.3398 0.0058 Crl transcriptional regulator<br />
csgA b1042 1.1757 0.0491 curlin, major subunit<br />
csgD b1040 0.3335 0.0298 CsgD transcriptional activator<br />
csgE b1039 0.3294 0.0116 curli production assembly/transport component<br />
csgG b1037 0.6373 0.0467 curli production component<br />
cspA b3556 0.1595 0.0272 cold shock protein CspA<br />
cspB b1557 0.0804 0.0020 cold shock protein CspB<br />
cspF b1558 0.1563 0.0010 cold shock protein CspF<br />
cspI b1552 0.4222 0.0364 cold shock protein CspI<br />
cvpA b2313 0.4563 0.0323 membrane protein required for colicin V production<br />
cyaY b3807 0.7207 0.0274 iron-binding frataxin homolog<br />
cybB b1418 0.4149 0.0121 cytochrome b561<br />
cybC b4236 0.5877 0.0172 cytochrome b562 (soluble)<br />
cynR b0338 1.4730 0.0246 CynR-Cyanate transcriptional activator<br />
cyoA b0432 1.6203 0.0356 cytochrome bo terminal oxidase subunit II<br />
cyoD b0429 1.2541 0.0388 cytochrome bo terminal oxidase subunit IV<br />
cyoE b0428 1.7729 0.0214 heme O synthase<br />
cysJ b2764 0.8120 0.0324 sulfite reductase flavoprotein subunit<br />
cysM b2421 0.7167 0.0338 O-acetylserine (thiol)-lyase B<br />
cytR b3934 0.3074 0.0076 CytR-cytidine<br />
dapA b2478 0.5629 0.0328 dihydrodipicolinate synthase<br />
dapF b3809 0.5061 0.0428 diaminopimelate epimerase<br />
dcm b1961 0.8273 0.0436 DNA cytosine methylase<br />
dcrB b3472 2.0849 0.0233 conserved protein involved in bacteriophage adsorption<br />
ddpA b1487 1.5696 0.0361 YddS<br />
ddpC b1485 2.0675 0.0310 YddQ<br />
degS b3235 0.4723 0.0130<br />
inner membrane serine protease required for the extracytoplasmic stress response<br />
mediated <strong>by</strong> sigmaE<br />
dfp b3639 0.4912 0.0038 P-pantothenate cysteine ligase / P-pantothenoylcysteine decarboxylase<br />
dnaB b4052 0.2288 0.0030 chromosome replication; chain elongation; part <strong>of</strong> primosome<br />
dnaG b3066 2.0680 0.0024 DNA biosynthesis; DNA primase<br />
dppC b3542 1.7014 0.0194 dipeptide ABC transporter<br />
dppF b3540 1.6050 0.0225 dipeptide ABC transporter<br />
dsbB b1185 0.7021 0.0043 dsbB protein<br />
dsbC b2893 0.5304 0.0070 DsbCoxidized<br />
dtd b3887 0.6855 0.0317 D-Tyr-tRNATyr deacylase<br />
dxr b0173 1.5005 0.0195 DXP reductoisomerase<br />
eda b1850 1.6768 0.0060<br />
oxaloacetate decarboxylase / 2-keto-3-deoxy-6-phosphogluconate aldolase / 2-<br />
keto-4-hydroxyglutarate aldolase<br />
elaA b2267 0.7645 0.0471 hypothetical protein<br />
elaC b2268 0.6841 0.0220 binuclear zinc phosphodiesterase monomer<br />
emrE b0543 1.4396 0.0395 EmrE SMR transporter<br />
entD b0583 0.7069 0.0132 phosphopantetheinyl transferase<br />
entE b0594 0.5193 0.0155 enterobactin synthase multienzyme complex<br />
envR b3264 0.6510 0.0447 EnvR transcriptional regulator<br />
epd b2927 0.4460 0.0015 erythrose 4-phosphate dehydrogenase<br />
eutN b2456 1.0929 0.0134 putative detox protein, ethanolamine utilization<br />
138