Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
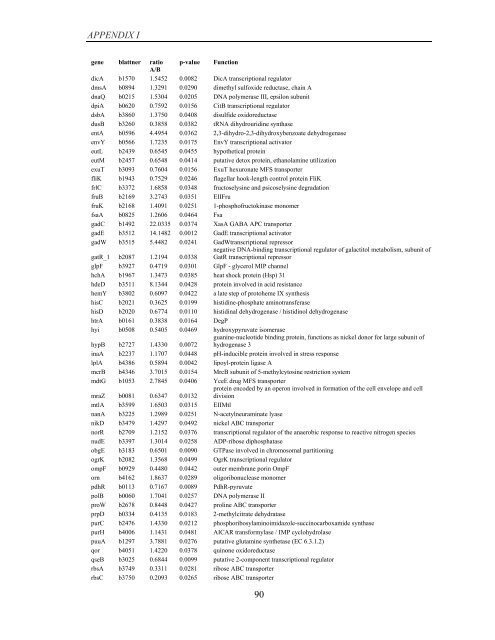
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
dicA b1570 1.5452 0.0082 DicA transcriptional regulator<br />
dmsA b0894 1.3291 0.0290 dimethyl sulfoxide reductase, chain A<br />
dnaQ b0215 1.5304 0.0205 DNA polymerase III, epsilon subunit<br />
dpiA b0620 0.7592 0.0156 CitB transcriptional regulator<br />
dsbA b3860 1.3750 0.0408 disulfide oxidoreductase<br />
dusB b3260 0.3858 0.0382 tRNA dihydrouridine synthase<br />
entA b0596 4.4954 0.0362 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase<br />
envY b0566 1.7235 0.0175 EnvY transcriptional activator<br />
eutL b2439 0.6545 0.0455 hypothetical protein<br />
eutM b2457 0.6548 0.0414 putative detox protein, ethanolamine utilization<br />
exuT b3093 0.7604 0.0156 ExuT hexuronate MFS transporter<br />
fliK b1943 0.7529 0.0246 flagellar hook-length control protein FliK<br />
frlC b3372 1.6858 0.0348 fructoselysine and psicoselysine degradation<br />
fruB b2169 3.2743 0.0351 EIIFru<br />
fruK b2168 1.4091 0.0251 1-phosph<strong>of</strong>ructokinase monomer<br />
fsaA b0825 1.2606 0.0464 Fsa<br />
gadC b1492 22.0335 0.0374 XasA GABA APC transporter<br />
gadE b3512 14.1482 0.0012 GadE transcriptional activator<br />
gadW b3515 5.4482 0.0241 GadWtranscriptional repressor<br />
gatR_1 b2087 1.2194 0.0338<br />
negative DNA-binding transcriptional regulator <strong>of</strong> galactitol metabolism, subunit <strong>of</strong><br />
GatR transcriptional repressor<br />
glpF b3927 0.4719 0.0301 GlpF - glycerol MIP channel<br />
hchA b1967 1.3473 0.0385 heat shock protein (Hsp) 31<br />
hdeD b3511 8.1344 0.0428 protein involved in acid resistance<br />
hemY b3802 0.6097 0.0422 a late step <strong>of</strong> protoheme IX synthesis<br />
hisC b2021 0.3625 0.0199 histidine-phosphate aminotransferase<br />
hisD b2020 0.6774 0.0110 histidinal dehydrogenase / histidinol dehydrogenase<br />
htrA b0161 0.3838 0.0164 DegP<br />
hyi b0508 0.5405 0.0469 hydroxypyruvate isomerase<br />
hypB b2727 1.4330 0.0072<br />
guanine-nucleotide binding protein, functions as nickel donor for large subunit <strong>of</strong><br />
hydrogenase 3<br />
inaA b2237 1.1707 0.0448 pH-inducible protein involved in stress response<br />
lplA b4386 0.5894 0.0042 lipoyl-protein ligase A<br />
mcrB b4346 3.7015 0.0154 MrcB subunit <strong>of</strong> 5-methylcytosine restriction system<br />
mdtG b1053 2.7845 0.0406 YceE drug MFS transporter<br />
mraZ b0081 0.6347 0.0132<br />
protein encoded <strong>by</strong> an operon involved in formation <strong>of</strong> the cell envelope and cell<br />
division<br />
mtlA b3599 1.6503 0.0315 EIIMtl<br />
nanA b3225 1.2989 0.0251 N-acetylneuraminate lyase<br />
nikD b3479 1.4297 0.0492 nickel ABC transporter<br />
norR b2709 1.2152 0.0376 transcriptional regulator <strong>of</strong> the anaerobic response to reactive nitrogen species<br />
nudE b3397 1.3014 0.0258 ADP-ribose diphosphatase<br />
obgE b3183 0.6501 0.0090 GTPase involved in chromosomal partitioning<br />
ogrK b2082 1.3568 0.0499 OgrK transcriptional regulator<br />
ompF b0929 0.4480 0.0442 outer membrane porin OmpF<br />
orn b4162 1.8637 0.0289 oligoribonuclease monomer<br />
pdhR b0113 0.7167 0.0089 PdhR-pyruvate<br />
polB b0060 1.7041 0.0257 DNA polymerase II<br />
proW b2678 0.8448 0.0427 proline ABC transporter<br />
prpD b0334 0.4135 0.0183 2-methylcitrate dehydratase<br />
purC b2476 1.4330 0.0212 phosphoribosylaminoimidazole-succinocarboxamide synthase<br />
purH b4006 1.1431 0.0481 AICAR transformylase / IMP cyclohydrolase<br />
puuA b1297 3.7881 0.0276 putative glutamine synthetase (EC 6.3.1.2)<br />
qor b4051 1.4220 0.0378 quinone oxidoreductase<br />
qseB b3025 0.6844 0.0099 putative 2-component transcriptional regulator<br />
rbsA b3749 0.3311 0.0281 ribose ABC transporter<br />
rbsC b3750 0.2093 0.0265 ribose ABC transporter<br />
90