Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
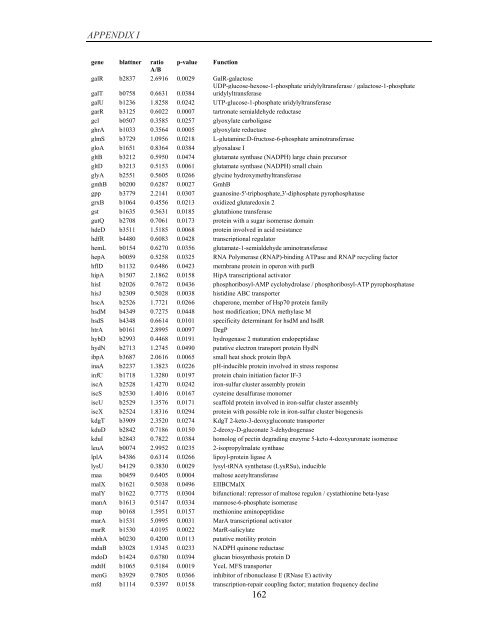
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
galR b2837 2.6916 0.0029 GalR-galactose<br />
galT b0758 0.6631 0.0384<br />
UDP-glucose-hexose-1-phosphate uridylyltransferase / galactose-1-phosphate<br />
uridylyltransferase<br />
galU b1236 1.8258 0.0242 UTP-glucose-1-phosphate uridylyltransferase<br />
garR b3125 0.6022 0.0007 tartronate semialdehyde reductase<br />
gcl b0507 0.3585 0.0257 glyoxylate carboligase<br />
ghrA b1033 0.3564 0.0005 glyoxylate reductase<br />
glmS b3729 1.0956 0.0218 L-glutamine:D-fructose-6-phosphate aminotransferase<br />
gloA b1651 0.8364 0.0384 glyoxalase I<br />
gltB b3212 0.5950 0.0474 glutamate synthase (NADPH) large chain precursor<br />
gltD b3213 0.5153 0.0061 glutamate synthase (NADPH) small chain<br />
glyA b2551 0.5605 0.0266 glycine hydroxymethyltransferase<br />
gmhB b0200 0.6287 0.0027 GmhB<br />
gpp b3779 2.2141 0.0307 guanosine-5'-triphosphate,3'-diphosphate pyrophosphatase<br />
grxB b1064 0.4556 0.0213 oxidized glutaredoxin 2<br />
gst b1635 0.5631 0.0185 glutathione transferase<br />
gutQ b2708 0.7061 0.0173 protein with a sugar isomerase domain<br />
hdeD b3511 1.5185 0.0068 protein involved in acid resistance<br />
hdfR b4480 0.6083 0.0428 transcriptional regulator<br />
hemL b0154 0.6270 0.0356 glutamate-1-semialdehyde aminotransferase<br />
hepA b0059 0.5258 0.0325 RNA Polymerase (RNAP)-binding ATPase and RNAP recycling factor<br />
hflD b1132 0.6486 0.0423 membrane protein in operon with purB<br />
hipA b1507 2.1862 0.0158 HipA transcriptional activator<br />
hisI b2026 0.7672 0.0436 phosphoribosyl-AMP cyclohydrolase / phosphoribosyl-ATP pyrophosphatase<br />
hisJ b2309 0.5028 0.0038 histidine ABC transporter<br />
hscA b2526 1.7721 0.0266 chaperone, member <strong>of</strong> Hsp70 protein family<br />
hsdM b4349 0.7275 0.0448 host modification; DNA methylase M<br />
hsdS b4348 0.6614 0.0101 specificity determinant for hsdM and hsdR<br />
htrA b0161 2.8995 0.0097 DegP<br />
hybD b2993 0.4468 0.0191 hydrogenase 2 maturation endopeptidase<br />
hydN b2713 1.2745 0.0490 putative electron transport protein HydN<br />
ibpA b3687 2.0616 0.0065 small heat shock protein IbpA<br />
inaA b2237 1.3823 0.0226 pH-inducible protein involved in stress response<br />
infC b1718 1.3280 0.0197 protein chain initiation factor IF-3<br />
iscA b2528 1.4270 0.0242 iron-sulfur cluster assembly protein<br />
iscS b2530 1.4016 0.0167 cysteine desulfurase monomer<br />
iscU b2529 1.3576 0.0171 scaffold protein involved in iron-sulfur cluster assembly<br />
iscX b2524 1.8316 0.0294 protein with possible role in iron-sulfur cluster bio<strong>gene</strong>sis<br />
kdgT b3909 2.3520 0.0274 KdgT 2-keto-3-deoxygluconate transporter<br />
kduD b2842 0.7186 0.0150 2-deoxy-D-gluconate 3-dehydrogenase<br />
kduI b2843 0.7822 0.0384 homolog <strong>of</strong> pectin degrading enzyme 5-keto 4-deoxyuronate isomerase<br />
leuA b0074 2.9952 0.0235 2-isopropylmalate synthase<br />
lplA b4386 0.6314 0.0266 lipoyl-protein ligase A<br />
lysU b4129 0.3830 0.0029 lysyl-tRNA synthetase (LysRSu), inducible<br />
maa b0459 0.6405 0.0004 maltose acetyltransferase<br />
malX b1621 0.5038 0.0496 EIIBCMalX<br />
malY b1622 0.7775 0.0304 bifunctional: repressor <strong>of</strong> maltose regulon / cystathionine beta-lyase<br />
manA b1613 0.5147 0.0334 mannose-6-phosphate isomerase<br />
map b0168 1.5951 0.0157 methionine aminopeptidase<br />
marA b1531 5.0995 0.0031 MarA transcriptional activator<br />
marR b1530 4.0195 0.0022 MarR-salicylate<br />
mbhA b0230 0.4200 0.0113 putative motility protein<br />
mdaB b3028 1.9345 0.0233 NADPH quinone reductase<br />
mdoD b1424 0.6780 0.0394 glucan biosynthesis protein D<br />
mdtH b1065 0.5184 0.0019 YceL MFS transporter<br />
menG b3929 0.7805 0.0366 inhibitor <strong>of</strong> ribonuclease E (RNase E) activity<br />
mfd b1114 0.5397 0.0158 transcription-repair coupling factor; mutation frequency decline<br />
162