Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
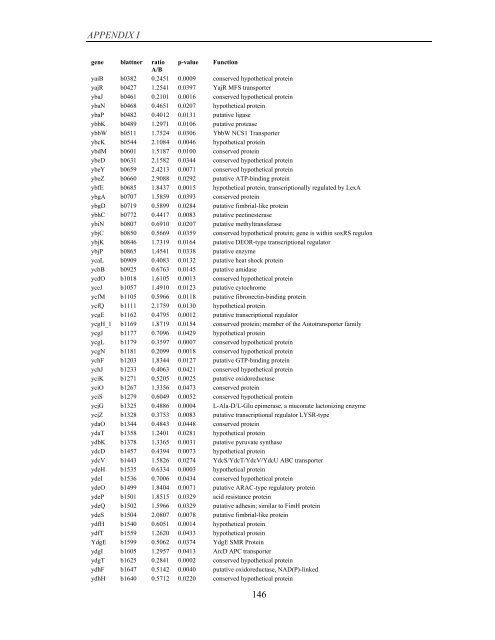
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
yaiB b0382 0.2451 0.0009 conserved hypothetical protein<br />
yajR b0427 1.2541 0.0397 YajR MFS transporter<br />
ybaJ b0461 0.2101 0.0016 conserved hypothetical protein<br />
ybaN b0468 0.4651 0.0207 hypothetical protein<br />
ybaP b0482 0.4012 0.0131 putative ligase<br />
ybbK b0489 1.2971 0.0106 putative protease<br />
ybbW b0511 1.7524 0.0306 YbbW NCS1 Transporter<br />
ybcK b0544 2.1084 0.0046 hypothetical protein<br />
ybdM b0601 1.5187 0.0100 conserved protein<br />
ybeD b0631 2.1582 0.0344 conserved hypothetical protein<br />
ybeY b0659 2.4213 0.0071 conserved hypothetical protein<br />
ybeZ b0660 2.9088 0.0292 putative ATP-binding protein<br />
ybfE b0685 1.8437 0.0015 hypothetical protein, transcriptionally regulated <strong>by</strong> LexA<br />
ybgA b0707 1.5859 0.0393 conserved protein<br />
ybgD b0719 0.5899 0.0284 putative fimbrial-like protein<br />
ybhC b0772 0.4417 0.0083 putative pectinesterase<br />
ybiN b0807 0.6910 0.0207 putative methyltransferase<br />
ybjC b0850 0.5669 0.0359 conserved hypothetical protein; <strong>gene</strong> is within soxRS regulon<br />
ybjK b0846 1.7319 0.0164 putative DEOR-type transcriptional regulator<br />
ybjP b0865 1.4541 0.0338 putative enzyme<br />
ycaL b0909 0.4083 0.0132 putative heat shock protein<br />
ycbB b0925 0.6763 0.0145 putative amidase<br />
ycdO b1018 1.6105 0.0013 conserved hypothetical protein<br />
yceJ b1057 1.4910 0.0123 putative cytochrome<br />
ycfM b1105 0.5966 0.0118 putative fibronectin-binding protein<br />
ycfQ b1111 2.1759 0.0130 hypothetical protein<br />
ycgE b1162 0.4795 0.0012 putative transcriptional regulator<br />
ycgH_1 b1169 1.8719 0.0154 conserved protein; member <strong>of</strong> the Autotransporter family<br />
ycgJ b1177 0.7096 0.0429 hypothetical protein<br />
ycgL b1179 0.3597 0.0007 conserved hypothetical protein<br />
ycgN b1181 0.2099 0.0018 conserved hypothetical protein<br />
ychF b1203 1.8344 0.0127 putative GTP-binding protein<br />
ychJ b1233 0.4063 0.0421 conserved hypothetical protein<br />
yciK b1271 0.5205 0.0025 putative oxidoreductase<br />
yciO b1267 1.3356 0.0473 conserved protein<br />
yciS b1279 0.6049 0.0052 conserved hypothetical protein<br />
ycjG b1325 0.4886 0.0004 L-Ala-D/L-Glu epimerase, a muconate lactonizing enzyme<br />
ycjZ b1328 0.3753 0.0083 putative transcriptional regulator LYSR-type<br />
ydaO b1344 0.4843 0.0448 conserved protein<br />
ydaT b1358 1.2401 0.0281 hypothetical protein<br />
ydbK b1378 1.3365 0.0031 putative pyruvate synthase<br />
ydcD b1457 0.4394 0.0073 hypothetical protein<br />
ydcV b1443 1.5826 0.0274 YdcS/YdcT/YdcV/YdcU ABC transporter<br />
ydeH b1535 0.6334 0.0003 hypothetical protein<br />
ydeI b1536 0.7006 0.0434 conserved hypothetical protein<br />
ydeO b1499 1.8404 0.0071 putative ARAC-type regulatory protein<br />
ydeP b1501 1.8515 0.0329 acid resistance protein<br />
ydeQ b1502 1.5966 0.0329 putative adhesin; similar to FimH protein<br />
ydeS b1504 2.0807 0.0078 putative fimbrial-like protein<br />
ydfH b1540 0.6051 0.0014 hypothetical protein<br />
ydfT b1559 1.2620 0.0433 hypothetical protein<br />
YdgE b1599 0.5062 0.0374 YdgE SMR Protein<br />
ydgI b1605 1.2957 0.0413 ArcD APC transporter<br />
ydgT b1625 0.2841 0.0002 conserved hypothetical protein<br />
ydhF b1647 0.5142 0.0040 putative oxidoreductase, NAD(P)-linked<br />
ydhH b1640 0.5712 0.0220 conserved hypothetical protein<br />
146