Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
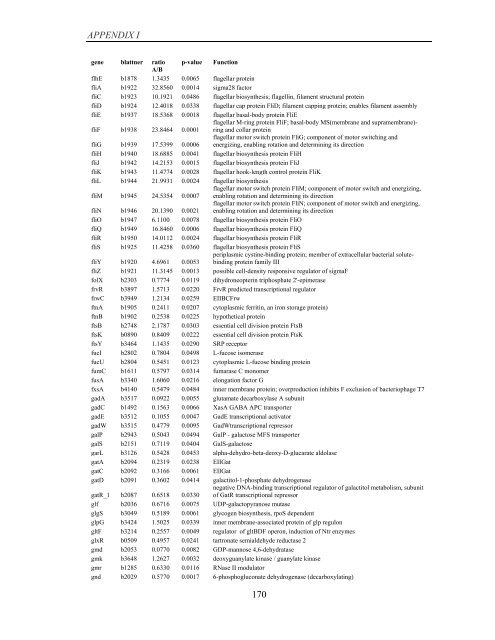
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
flhE b1878 1.3435 0.0065 flagellar protein<br />
fliA b1922 32.8560 0.0014 sigma28 factor<br />
fliC b1923 10.1921 0.0486 flagellar biosynthesis; flagellin, filament structural protein<br />
fliD b1924 12.4018 0.0338 flagellar cap protein FliD; filament capping protein; enables filament assembly<br />
fliE b1937 18.5368 0.0018 flagellar basal-body protein FliE<br />
fliF b1938 23.8464 0.0001<br />
flagellar M-ring protein FliF; basal-body MS(membrane and supramembrane)-<br />
ring and collar protein<br />
fliG b1939 17.5399 0.0006<br />
flagellar motor switch protein FliG; component <strong>of</strong> motor switching and<br />
energizing, enabling rotation and determining its direction<br />
fliH b1940 18.6885 0.0041 flagellar biosynthesis protein FliH<br />
fliJ b1942 14.2153 0.0015 flagellar biosynthesis protein FliJ<br />
fliK b1943 11.4774 0.0028 flagellar hook-length control protein FliK<br />
fliL b1944 21.9931 0.0024 flagellar biosynthesis<br />
fliM b1945 24.5354 0.0007<br />
flagellar motor switch protein FliM; component <strong>of</strong> motor switch and energizing,<br />
enabling rotation and determining its direction<br />
fliN b1946 20.1390 0.0021<br />
flagellar motor switch protein FliN; component <strong>of</strong> motor switch and energizing,<br />
enabling rotation and determining its direction<br />
fliO b1947 6.1100 0.0078 flagellar biosynthesis protein FliO<br />
fliQ b1949 16.8460 0.0006 flagellar biosynthesis protein FliQ<br />
fliR b1950 14.0112 0.0024 flagellar biosynthesis protein FliR<br />
fliS b1925 11.4258 0.0360 flagellar biosynthesis protein FliS<br />
fliY b1920 4.6961 0.0053<br />
periplasmic cystine-binding protein; member <strong>of</strong> extracellular bacterial solutebinding<br />
protein family III<br />
fliZ b1921 11.3145 0.0013 possible cell-density responsive regulator <strong>of</strong> sigmaF<br />
folX b2303 0.7774 0.0119 dihydroneopterin triphosphate 2'-epimerase<br />
frvR b3897 1.5713 0.0220 FrvR predicted transcriptional regulator<br />
frwC b3949 1.2134 0.0259 EIIBCFrw<br />
ftnA b1905 0.2411 0.0207 cytoplasmic ferritin, an iron storage protein)<br />
ftnB b1902 0.2538 0.0225 hypothetical protein<br />
ftsB b2748 2.1787 0.0303 essential cell division protein FtsB<br />
ftsK b0890 0.8409 0.0222 essential cell division protein FtsK<br />
ftsY b3464 1.1435 0.0290 SRP receptor<br />
fucI b2802 0.7804 0.0498 L-fucose isomerase<br />
fucU b2804 0.5451 0.0123 cytoplasmic L-fucose binding protein<br />
fumC b1611 0.5797 0.0314 fumarase C monomer<br />
fusA b3340 1.6060 0.0216 elongation factor G<br />
fxsA b4140 0.5479 0.0484 inner membrane protein; overproduction inhibits F exclusion <strong>of</strong> bacteriophage T7<br />
gadA b3517 0.0922 0.0055 glutamate decarboxylase A subunit<br />
gadC b1492 0.1563 0.0066 XasA GABA APC transporter<br />
gadE b3512 0.1055 0.0047 GadE transcriptional activator<br />
gadW b3515 0.4779 0.0095 GadWtranscriptional repressor<br />
galP b2943 0.5043 0.0494 GalP - galactose MFS transporter<br />
galS b2151 0.7119 0.0404 GalS-galactose<br />
garL b3126 0.5428 0.0453 alpha-dehydro-beta-deoxy-D-glucarate aldolase<br />
gatA b2094 0.2319 0.0238 EIIGat<br />
gatC b2092 0.3166 0.0061 EIIGat<br />
gatD b2091 0.3602 0.0414 galactitol-1-phosphate dehydrogenase<br />
gatR_1 b2087 0.6518 0.0330<br />
negative DNA-binding transcriptional regulator <strong>of</strong> galactitol metabolism, subunit<br />
<strong>of</strong> GatR transcriptional repressor<br />
glf b2036 0.6716 0.0075 UDP-galactopyranose mutase<br />
glgS b3049 0.5189 0.0061 glycogen biosynthesis, rpoS dependent<br />
glpG b3424 1.5025 0.0339 inner membrane-associated protein <strong>of</strong> glp regulon<br />
gltF b3214 0.2557 0.0049 regulator <strong>of</strong> gltBDF operon, induction <strong>of</strong> Ntr enzymes<br />
glxR b0509 0.4957 0.0241 tartronate semialdehyde reductase 2<br />
gmd b2053 0.0770 0.0082 GDP-mannose 4,6-dehydratase<br />
gmk b3648 1.2627 0.0032 deoxyguanylate kinase / guanylate kinase<br />
gmr b1285 0.6330 0.0116 RNase II modulator<br />
gnd b2029 0.5770 0.0017 6-phosphogluconate dehydrogenase (decarboxylating)<br />
170