Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
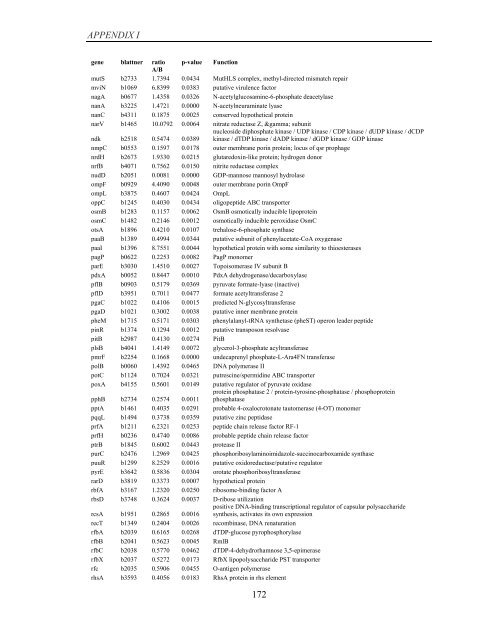
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
mutS b2733 1.7394 0.0434 MutHLS complex, methyl-directed mismatch repair<br />
mviN b1069 6.8399 0.0383 putative virulence factor<br />
nagA b0677 1.4358 0.0326 N-acetylglucosamine-6-phosphate deacetylase<br />
nanA b3225 1.4721 0.0000 N-acetylneuraminate lyase<br />
nanC b4311 0.1875 0.0025 conserved hypothetical protein<br />
narV b1465 10.0792 0.0064 nitrate reductase Z, γ subunit<br />
ndk b2518 0.5474 0.0389<br />
nucleoside diphosphate kinase / UDP kinase / CDP kinase / dUDP kinase / dCDP<br />
kinase / dTDP kinase / dADP kinase / dGDP kinase / GDP kinase<br />
nmpC b0553 0.1597 0.0178 outer membrane porin protein; locus <strong>of</strong> qsr prophage<br />
nrdH b2673 1.9330 0.0215 glutaredoxin-like protein; hydrogen donor<br />
nrfB b4071 0.7562 0.0150 nitrite reductase complex<br />
nudD b2051 0.0081 0.0000 GDP-mannose mannosyl hydrolase<br />
ompF b0929 4.4090 0.0048 outer membrane porin OmpF<br />
ompL b3875 0.4607 0.0424 OmpL<br />
oppC b1245 0.4030 0.0434 oligopeptide ABC transporter<br />
osmB b1283 0.1157 0.0062 OsmB osmotically inducible lipoprotein<br />
osmC b1482 0.2146 0.0012 osmotically inducible peroxidase OsmC<br />
otsA b1896 0.4210 0.0107 trehalose-6-phosphate synthase<br />
paaB b1389 0.4994 0.0344 putative subunit <strong>of</strong> phenylacetate-CoA oxygenase<br />
paaI b1396 8.7551 0.0044 hypothetical protein with some similarity to thioesterases<br />
pagP b0622 0.2253 0.0082 PagP monomer<br />
parE b3030 1.4510 0.0027 Topoisomerase IV subunit B<br />
pdxA b0052 0.8447 0.0010 PdxA dehydrogenase/decarboxylase<br />
pflB b0903 0.5179 0.0369 pyruvate formate-lyase (inactive)<br />
pflD b3951 0.7011 0.0477 formate acetyltransferase 2<br />
pgaC b1022 0.4106 0.0015 predicted N-glycosyltransferase<br />
pgaD b1021 0.3002 0.0038 putative inner membrane protein<br />
pheM b1715 0.5171 0.0303 phenylalanyl-tRNA synthetase (pheST) operon leader peptide<br />
pinR b1374 0.1294 0.0012 putative transposon resolvase<br />
pitB b2987 0.4130 0.0274 PitB<br />
plsB b4041 1.4149 0.0072 glycerol-3-phosphate acyltransferase<br />
pmrF b2254 0.1668 0.0000 undecaprenyl phosphate-L-Ara4FN transferase<br />
polB b0060 1.4392 0.0465 DNA polymerase II<br />
potC b1124 0.7024 0.0321 putrescine/spermidine ABC transporter<br />
poxA b4155 0.5601 0.0149 putative regulator <strong>of</strong> pyruvate oxidase<br />
pphB b2734 0.2574 0.0011<br />
protein phosphatase 2 / protein-tyrosine-phosphatase / phosphoprotein<br />
phosphatase<br />
pptA b1461 0.4035 0.0291 probable 4-oxalocrotonate tautomerase (4-OT) monomer<br />
pqqL b1494 0.3738 0.0359 putative zinc peptidase<br />
prfA b1211 6.2321 0.0253 peptide chain release factor RF-1<br />
prfH b0236 0.4740 0.0086 probable peptide chain release factor<br />
ptrB b1845 0.6002 0.0443 protease II<br />
purC b2476 1.2969 0.0425 phosphoribosylaminoimidazole-succinocarboxamide synthase<br />
puuR b1299 8.2529 0.0016 putative oxidoreductase/putative regulator<br />
pyrE b3642 0.5836 0.0304 orotate phosphoribosyltransferase<br />
rarD b3819 0.3373 0.0007 hypothetical protein<br />
rbfA b3167 1.2320 0.0250 ribosome-binding factor A<br />
rbsD b3748 0.3624 0.0037 D-ribose utilization<br />
rcsA b1951 0.2865 0.0016<br />
positive DNA-binding transcriptional regulator <strong>of</strong> capsular polysaccharide<br />
synthesis, activates its own <strong>expression</strong><br />
recT b1349 0.2404 0.0026 recombinase, DNA renaturation<br />
rfbA b2039 0.6165 0.0268 dTDP-glucose pyrophosphorylase<br />
rfbB b2041 0.5623 0.0045 RmlB<br />
rfbC b2038 0.5770 0.0462 dTDP-4-dehydrorhamnose 3,5-epimerase<br />
rfbX b2037 0.5272 0.0173 RfbX lipopolysaccharide PST transporter<br />
rfc b2035 0.5906 0.0455 O-antigen polymerase<br />
rhsA b3593 0.4056 0.0183 RhsA protein in rhs element<br />
172