Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
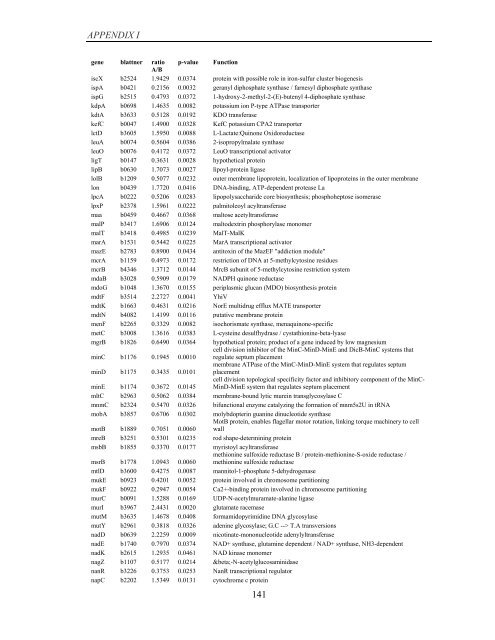
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
iscX b2524 1.9429 0.0374 protein with possible role in iron-sulfur cluster bio<strong>gene</strong>sis<br />
ispA b0421 0.2156 0.0032 geranyl diphosphate synthase / farnesyl diphosphate synthase<br />
ispG b2515 0.4793 0.0372 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase<br />
kdpA b0698 1.4635 0.0082 potassium ion P-type ATPase transporter<br />
kdtA b3633 0.5128 0.0192 KDO transferase<br />
kefC b0047 1.4900 0.0328 KefC potassium CPA2 transporter<br />
lctD b3605 1.5950 0.0088 L-Lactate:Quinone Oxidoreductase<br />
leuA b0074 0.5604 0.0386 2-isopropylmalate synthase<br />
leuO b0076 0.4172 0.0372 LeuO transcriptional activator<br />
ligT b0147 0.3631 0.0028 hypothetical protein<br />
lipB b0630 1.7073 0.0027 lipoyl-protein ligase<br />
lolB b1209 0.5077 0.0232 outer membrane lipoprotein, localization <strong>of</strong> lipoproteins in the outer membrane<br />
lon b0439 1.7720 0.0416 DNA-binding, ATP-dependent protease La<br />
lpcA b0222 0.5206 0.0283 lipopolysaccharide core biosynthesis; phosphoheptose isomerase<br />
lpxP b2378 1.5961 0.0222 palmitoleoyl acyltransferase<br />
maa b0459 0.4667 0.0368 maltose acetyltransferase<br />
malP b3417 1.6906 0.0124 maltodextrin phosphorylase monomer<br />
malT b3418 0.4985 0.0239 MalT-MalK<br />
marA b1531 0.5442 0.0225 MarA transcriptional activator<br />
mazE b2783 0.8900 0.0434 antitoxin <strong>of</strong> the MazEF "addiction module"<br />
mcrA b1159 0.4973 0.0172 restriction <strong>of</strong> DNA at 5-methylcytosine residues<br />
mcrB b4346 1.3712 0.0144 MrcB subunit <strong>of</strong> 5-methylcytosine restriction system<br />
mdaB b3028 0.5909 0.0179 NADPH quinone reductase<br />
mdoG b1048 1.3670 0.0155 periplasmic glucan (MDO) biosynthesis protein<br />
mdtF b3514 2.2727 0.0041 YhiV<br />
mdtK b1663 0.4631 0.0216 NorE multidrug efflux MATE transporter<br />
mdtN b4082 1.4199 0.0116 putative membrane protein<br />
menF b2265 0.3329 0.0082 isochorismate synthase, menaquinone-specific<br />
metC b3008 1.3616 0.0383 L-cysteine desulfhydrase / cystathionine-beta-lyase<br />
mgrB b1826 0.6490 0.0364 hypothetical protein; product <strong>of</strong> a <strong>gene</strong> induced <strong>by</strong> low magnesium<br />
minC b1176 0.1945 0.0010<br />
cell division inhibitor <strong>of</strong> the MinC-MinD-MinE and DicB-MinC systems that<br />
regulate septum placement<br />
minD b1175 0.3435 0.0101<br />
membrane ATPase <strong>of</strong> the MinC-MinD-MinE system that regulates septum<br />
placement<br />
minE b1174 0.3672 0.0145<br />
cell division topological specificity factor and inhibitory component <strong>of</strong> the MinC-<br />
MinD-MinE system that regulates septum placement<br />
mltC b2963 0.5062 0.0384 membrane-bound lytic murein transglycosylase C<br />
mnmC b2324 0.5470 0.0326 bifunctional enzyme catalyzing the formation <strong>of</strong> mnm5s2U in tRNA<br />
mobA b3857 0.6706 0.0302 molybdopterin guanine dinucleotide synthase<br />
motB b1889 0.7051 0.0060<br />
MotB protein, enables flagellar motor rotation, linking torque machinery to cell<br />
wall<br />
mreB b3251 0.5301 0.0235 rod shape-determining protein<br />
msbB b1855 0.3370 0.0177 myristoyl acyltransferase<br />
msrB b1778 1.0943 0.0060<br />
methionine sulfoxide reductase B / protein-methionine-S-oxide reductase /<br />
methionine sulfoxide reductase<br />
mtlD b3600 0.4275 0.0087 mannitol-1-phosphate 5-dehydrogenase<br />
mukE b0923 0.4201 0.0052 protein involved in chromosome partitioning<br />
mukF b0922 0.2947 0.0054 Ca2+-binding protein involved in chromosome partitioning<br />
murC b0091 1.5288 0.0169 UDP-N-acetylmuramate-alanine ligase<br />
murI b3967 2.4431 0.0020 glutamate racemase<br />
mutM b3635 1.4678 0.0408 formamidopyrimidine DNA glycosylase<br />
mutY b2961 0.3818 0.0326 adenine glycosylase; G.C --> T.A transversions<br />
nadD b0639 2.2259 0.0009 nicotinate-mononucleotide adenylyltransferase<br />
nadE b1740 0.7970 0.0374 NAD+ synthase, glutamine dependent / NAD+ synthase, NH3-dependent<br />
nadK b2615 1.2935 0.0461 NAD kinase monomer<br />
nagZ b1107 0.5177 0.0214 β-N-acetylglucosaminidase<br />
nanR b3226 0.3753 0.0253 NanR transcriptional regulator<br />
napC b2202 1.5349 0.0131 cytochrome c protein<br />
141