Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
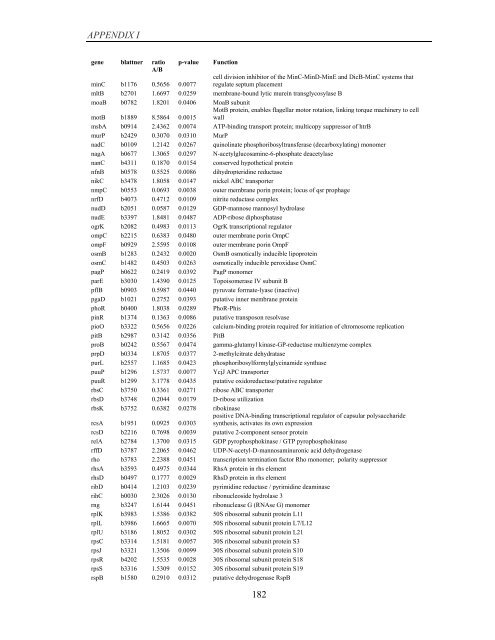
APPENDIX I<br />
<strong>gene</strong> blattner ratio<br />
A/B<br />
p-value<br />
Function<br />
cell division inhibitor <strong>of</strong> the MinC-MinD-MinE and DicB-MinC systems that<br />
regulate septum placement<br />
minC b1176 0.5656 0.0077<br />
mltB b2701 1.6697 0.0259 membrane-bound lytic murein transglycosylase B<br />
moaB b0782 1.8201 0.0406 MoaB subunit<br />
MotB protein, enables flagellar motor rotation, linking torque machinery to cell<br />
wall<br />
motB b1889 8.5864 0.0015<br />
msbA b0914 2.4362 0.0074 ATP-binding transport protein; multicopy suppressor <strong>of</strong> htrB<br />
murP b2429 0.3070 0.0310 MurP<br />
nadC b0109 1.2142 0.0267 quinolinate phosphoribosyltransferase (decarboxylating) monomer<br />
nagA b0677 1.3065 0.0297 N-acetylglucosamine-6-phosphate deacetylase<br />
nanC b4311 0.1870 0.0154 conserved hypothetical protein<br />
nfnB b0578 0.5525 0.0086 dihydropteridine reductase<br />
nikC b3478 1.8058 0.0147 nickel ABC transporter<br />
nmpC b0553 0.0693 0.0038 outer membrane porin protein; locus <strong>of</strong> qsr prophage<br />
nrfD b4073 0.4712 0.0109 nitrite reductase complex<br />
nudD b2051 0.0587 0.0129 GDP-mannose mannosyl hydrolase<br />
nudE b3397 1.8481 0.0487 ADP-ribose diphosphatase<br />
ogrK b2082 0.4983 0.0113 OgrK transcriptional regulator<br />
ompC b2215 0.6383 0.0480 outer membrane porin OmpC<br />
ompF b0929 2.5595 0.0108 outer membrane porin OmpF<br />
osmB b1283 0.2432 0.0020 OsmB osmotically inducible lipoprotein<br />
osmC b1482 0.4503 0.0263 osmotically inducible peroxidase OsmC<br />
pagP b0622 0.2419 0.0392 PagP monomer<br />
parE b3030 1.4390 0.0125 Topoisomerase IV subunit B<br />
pflB b0903 0.5987 0.0440 pyruvate formate-lyase (inactive)<br />
pgaD b1021 0.2752 0.0393 putative inner membrane protein<br />
phoR b0400 1.8038 0.0289 PhoR-Phis<br />
pinR b1374 0.1363 0.0086 putative transposon resolvase<br />
pioO b3322 0.5656 0.0226 calcium-binding protein required for initiation <strong>of</strong> chromosome replication<br />
pitB b2987 0.3142 0.0356 PitB<br />
proB b0242 0.5567 0.0474 gamma-glutamyl kinase-GP-reductase multienzyme complex<br />
prpD b0334 1.8705 0.0377 2-methylcitrate dehydratase<br />
purL b2557 1.1685 0.0423 phosphoribosylformylglycinamide synthase<br />
puuP b1296 1.5737 0.0077 YcjJ APC transporter<br />
puuR b1299 3.1778 0.0435 putative oxidoreductase/putative regulator<br />
rbsC b3750 0.3361 0.0271 ribose ABC transporter<br />
rbsD b3748 0.2044 0.0179 D-ribose utilization<br />
rbsK b3752 0.6382 0.0278 ribokinase<br />
positive DNA-binding transcriptional regulator <strong>of</strong> capsular polysaccharide<br />
rcsA b1951 0.0925 0.0303 synthesis, activates its own <strong>expression</strong><br />
rcsD b2216 0.7698 0.0039 putative 2-component sensor protein<br />
relA b2784 1.3700 0.0315 GDP pyrophosphokinase / GTP pyrophosphokinase<br />
rffD b3787 2.2065 0.0462 UDP-N-acetyl-D-mannosaminuronic acid dehydrogenase<br />
rho b3783 2.2388 0.0451 transcription termination factor Rho monomer; polarity suppressor<br />
rhsA b3593 0.4975 0.0344 RhsA protein in rhs element<br />
rhsD b0497 0.1777 0.0029 RhsD protein in rhs element<br />
ribD b0414 1.2103 0.0239 pyrimidine reductase / pyrimidine deaminase<br />
rihC b0030 2.3026 0.0130 ribonucleoside hydrolase 3<br />
rng b3247 1.6144 0.0451 ribonuclease G (RNAse G) monomer<br />
rplK b3983 1.5386 0.0382 50S ribosomal subunit protein L11<br />
rplL b3986 1.6665 0.0070 50S ribosomal subunit protein L7/L12<br />
rplU b3186 1.8052 0.0302 50S ribosomal subunit protein L21<br />
rpsC b3314 1.5181 0.0057 30S ribosomal subunit protein S3<br />
rpsJ b3321 1.3506 0.0099 30S ribosomal subunit protein S10<br />
rpsR b4202 1.5535 0.0028 30S ribosomal subunit protein S18<br />
rpsS b3316 1.5309 0.0152 30S ribosomal subunit protein S19<br />
rspB b1580 0.2910 0.0312 putative dehydrogenase RspB<br />
182