Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
APPENDIX I<br />
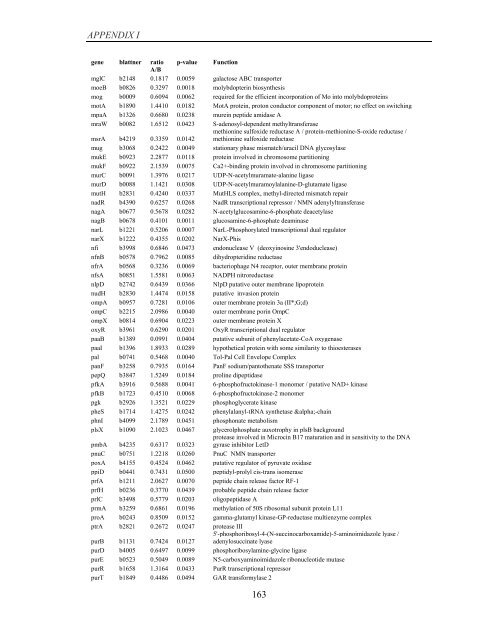
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
mglC b2148 0.1817 0.0059 galactose ABC transporter<br />
moeB b0826 0.3297 0.0018 molybdopterin biosynthesis<br />
mog b0009 0.6094 0.0062 required for the efficient incorporation <strong>of</strong> Mo into molybdoproteins<br />
motA b1890 1.4410 0.0182 MotA protein, proton conductor component <strong>of</strong> motor; no effect on switching<br />
mpaA b1326 0.6680 0.0238 murein peptide amidase A<br />
mraW b0082 1.6512 0.0423 S-adenosyl-dependent methyltransferase<br />
msrA b4219 0.3359 0.0142<br />
methionine sulfoxide reductase A / protein-methionine-S-oxide reductase /<br />
methionine sulfoxide reductase<br />
mug b3068 0.2422 0.0049 stationary phase mismatch/uracil DNA glycosylase<br />
mukE b0923 2.2877 0.0118 protein involved in chromosome partitioning<br />
mukF b0922 2.1539 0.0075 Ca2+-binding protein involved in chromosome partitioning<br />
murC b0091 1.3976 0.0217 UDP-N-acetylmuramate-alanine ligase<br />
murD b0088 1.1421 0.0308 UDP-N-acetylmuramoylalanine-D-glutamate ligase<br />
mutH b2831 0.4240 0.0337 MutHLS complex, methyl-directed mismatch repair<br />
nadR b4390 0.6257 0.0268 NadR transcriptional repressor / NMN adenylyltransferase<br />
nagA b0677 0.5678 0.0282 N-acetylglucosamine-6-phosphate deacetylase<br />
nagB b0678 0.4101 0.0011 glucosamine-6-phosphate deaminase<br />
narL b1221 0.5206 0.0007 NarL-Phosphorylated transcriptional dual regulator<br />
narX b1222 0.4355 0.0202 NarX-Phis<br />
nfi b3998 0.6846 0.0473 endonuclease V (deoxyinosine 3'endoduclease)<br />
nfnB b0578 0.7962 0.0085 dihydropteridine reductase<br />
nfrA b0568 0.3236 0.0069 bacteriophage N4 receptor, outer membrane protein<br />
nfsA b0851 1.5581 0.0063 NADPH nitroreductase<br />
nlpD b2742 0.6439 0.0366 NlpD putative outer membrane lipoprotein<br />
nudH b2830 1.4474 0.0158 putative invasion protein<br />
ompA b0957 0.7281 0.0106 outer membrane protein 3a (II*;G;d)<br />
ompC b2215 2.0986 0.0040 outer membrane porin OmpC<br />
ompX b0814 0.6904 0.0223 outer membrane protein X<br />
oxyR b3961 0.6290 0.0201 OxyR transcriptional dual regulator<br />
paaB b1389 0.0991 0.0404 putative subunit <strong>of</strong> phenylacetate-CoA oxygenase<br />
paaI b1396 1.8933 0.0289 hypothetical protein with some similarity to thioesterases<br />
pal b0741 0.5468 0.0040 Tol-Pal Cell Envelope Complex<br />
panF b3258 0.7935 0.0164 PanF sodium/pantothenate SSS transporter<br />
pepQ b3847 1.5249 0.0184 proline dipeptidase<br />
pfkA b3916 0.5688 0.0041 6-phosph<strong>of</strong>ructokinase-1 monomer / putative NAD+ kinase<br />
pfkB b1723 0.4510 0.0068 6-phosph<strong>of</strong>ructokinase-2 monomer<br />
pgk b2926 1.3521 0.0229 phosphoglycerate kinase<br />
pheS b1714 1.4275 0.0242 phenylalanyl-tRNA synthetase α-chain<br />
phnI b4099 2.1789 0.0451 phosphonate metabolism<br />
plsX b1090 2.1023 0.0467 glycerolphosphate auxotrophy in plsB background<br />
pmbA b4235 0.6317 0.0323<br />
protease involved in Microcin B17 maturation and in sensitivity to the DNA<br />
gyrase inhibitor LetD<br />
pnuC b0751 1.2218 0.0260 PnuC NMN transporter<br />
poxA b4155 0.4524 0.0462 putative regulator <strong>of</strong> pyruvate oxidase<br />
ppiD b0441 0.7431 0.0500 peptidyl-prolyl cis-trans isomerase<br />
prfA b1211 2.0627 0.0070 peptide chain release factor RF-1<br />
prfH b0236 0.3770 0.0439 probable peptide chain release factor<br />
prlC b3498 0.5779 0.0203 oligopeptidase A<br />
prmA b3259 0.6861 0.0196 methylation <strong>of</strong> 50S ribosomal subunit protein L11<br />
proA b0243 0.8509 0.0152 gamma-glutamyl kinase-GP-reductase multienzyme complex<br />
ptrA b2821 0.2672 0.0247 protease III<br />
purB b1131 0.7424 0.0127<br />
5'-phosphoribosyl-4-(N-succinocarboxamide)-5-aminoimidazole lyase /<br />
adenylosuccinate lyase<br />
purD b4005 0.6497 0.0099 phosphoribosylamine-glycine ligase<br />
purE b0523 0.5049 0.0089 N5-carboxyaminoimidazole ribonucleotide mutase<br />
purR b1658 1.3164 0.0433 PurR transcriptional repressor<br />
purT b1849 0.4486 0.0494 GAR transformylase 2<br />
163