Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
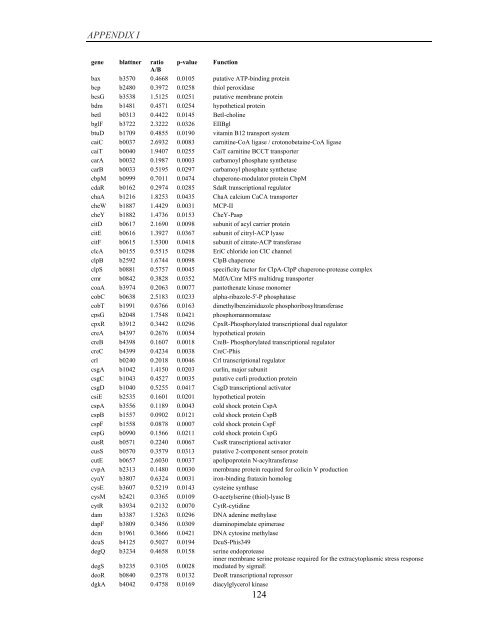
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
bax b3570 0.4668 0.0105 putative ATP-binding protein<br />
bcp b2480 0.3972 0.0258 thiol peroxidase<br />
bcsG b3538 1.5125 0.0251 putative membrane protein<br />
bdm b1481 0.4571 0.0254 hypothetical protein<br />
betI b0313 0.4422 0.0145 BetI-choline<br />
bglF b3722 2.3222 0.0326 EIIBgl<br />
btuD b1709 0.4855 0.0190 vitamin B12 transport system<br />
caiC b0037 2.6932 0.0083 carnitine-CoA ligase / crotonobetaine-CoA ligase<br />
caiT b0040 1.9407 0.0255 CaiT carnitine BCCT transporter<br />
carA b0032 0.1987 0.0003 carbamoyl phosphate synthetase<br />
carB b0033 0.5195 0.0297 carbamoyl phosphate synthetase<br />
cbpM b0999 0.7011 0.0474 chaperone-modulator protein CbpM<br />
cdaR b0162 0.2974 0.0285 SdaR transcriptional regulator<br />
chaA b1216 1.8253 0.0435 ChaA calcium CaCA transporter<br />
cheW b1887 1.4429 0.0031 MCP-II<br />
cheY b1882 1.4736 0.0153 CheY-Pasp<br />
citD b0617 2.1690 0.0098 subunit <strong>of</strong> acyl carrier protein<br />
citE b0616 1.3927 0.0367 subunit <strong>of</strong> citryl-ACP lyase<br />
citF b0615 1.5300 0.0418 subunit <strong>of</strong> citrate-ACP transferase<br />
clcA b0155 0.5515 0.0298 EriC chloride ion ClC channel<br />
clpB b2592 1.6744 0.0098 ClpB chaperone<br />
clpS b0881 0.5757 0.0045 specificity factor for ClpA-ClpP chaperone-protease complex<br />
cmr b0842 0.3828 0.0352 MdfA/Cmr MFS multidrug transporter<br />
coaA b3974 0.2063 0.0077 pantothenate kinase monomer<br />
cobC b0638 2.5183 0.0233 alpha-ribazole-5'-P phosphatase<br />
cobT b1991 0.6766 0.0163 dimethylbenzimidazole phosphoribosyltransferase<br />
cpsG b2048 1.7548 0.0421 phosphomannomutase<br />
cpxR b3912 0.3442 0.0296 CpxR-Phosphorylated transcriptional dual regulator<br />
creA b4397 0.2676 0.0054 hypothetical protein<br />
creB b4398 0.1607 0.0018 CreB- Phosphorylated transcriptional regulator<br />
creC b4399 0.4234 0.0038 CreC-Phis<br />
crl b0240 0.2018 0.0046 Crl transcriptional regulator<br />
csgA b1042 1.4150 0.0203 curlin, major subunit<br />
csgC b1043 0.4527 0.0035 putative curli production protein<br />
csgD b1040 0.5255 0.0417 CsgD transcriptional activator<br />
csiE b2535 0.1601 0.0201 hypothetical protein<br />
cspA b3556 0.1189 0.0043 cold shock protein CspA<br />
cspB b1557 0.0902 0.0121 cold shock protein CspB<br />
cspF b1558 0.0878 0.0007 cold shock protein CspF<br />
cspG b0990 0.1566 0.0211 cold shock protein CspG<br />
cusR b0571 0.2240 0.0067 CusR transcriptional activator<br />
cusS b0570 0.3579 0.0313 putative 2-component sensor protein<br />
cutE b0657 2.6030 0.0037 apolipoprotein N-acyltransferase<br />
cvpA b2313 0.1480 0.0030 membrane protein required for colicin V production<br />
cyaY b3807 0.6324 0.0031 iron-binding frataxin homolog<br />
cysE b3607 0.5219 0.0143 cysteine synthase<br />
cysM b2421 0.3365 0.0109 O-acetylserine (thiol)-lyase B<br />
cytR b3934 0.2132 0.0070 CytR-cytidine<br />
dam b3387 1.5263 0.0296 DNA adenine methylase<br />
dapF b3809 0.3456 0.0309 diaminopimelate epimerase<br />
dcm b1961 0.3666 0.0421 DNA cytosine methylase<br />
dcuS b4125 0.5027 0.0194 DcuS-Phis349<br />
degQ b3234 0.4658 0.0158 serine endoprotease<br />
degS b3235 0.3105 0.0028<br />
inner membrane serine protease required for the extracytoplasmic stress response<br />
mediated <strong>by</strong> sigmaE<br />
deoR b0840 0.2578 0.0132 DeoR transcriptional repressor<br />
dgkA b4042 0.4758 0.0169 diacylglycerol kinase<br />
124