Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
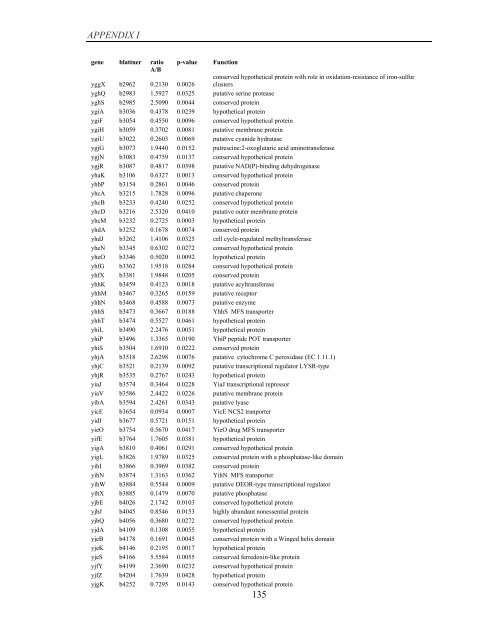
APPENDIX I<br />
<strong>gene</strong> blattner ratio<br />
A/B<br />
p-value<br />
yggX b2962 0.2130 0.0026<br />
Function<br />
conserved hypothetical protein with role in oxidation-resistance <strong>of</strong> iron-sulfur<br />
clusters<br />
yghQ b2983 1.5927 0.0325 putative serine protease<br />
yghS b2985 2.5090 0.0044 conserved protein<br />
ygiA b3036 0.4378 0.0239 hypothetical protein<br />
ygiF b3054 0.4550 0.0096 conserved hypothetical protein<br />
ygiH b3059 0.3702 0.0081 putative membrane protein<br />
ygiU b3022 0.2603 0.0069 putative cyanide hydratase<br />
ygjG b3073 1.9440 0.0152 putrescine:2-oxoglutaric acid aminotransferase<br />
ygjN b3083 0.4759 0.0137 conserved hypothetical protein<br />
ygjR b3087 0.4817 0.0398 putative NAD(P)-binding dehydrogenase<br />
yhaK b3106 0.6327 0.0013 conserved hypothetical protein<br />
yhbP b3154 0.2861 0.0046 conserved protein<br />
yhcA b3215 1.7828 0.0096 putative chaperone<br />
yhcB b3233 0.4240 0.0252 conserved hypothetical protein<br />
yhcD b3216 2.5320 0.0410 putative outer membrane protein<br />
yhcM b3232 0.2725 0.0003 hypothetical protein<br />
yhdA b3252 0.1678 0.0074 conserved protein<br />
yhdJ b3262 1.4106 0.0325 cell cycle-regulated methyltransferase<br />
yheN b3345 0.6302 0.0272 conserved hypothetical protein<br />
yheO b3346 0.5020 0.0092 hypothetical protein<br />
yhfG b3362 1.9518 0.0284 conserved hypothetical protein<br />
yhfX b3381 1.9848 0.0205 conserved protein<br />
yhhK b3459 0.4123 0.0018 putative acyltransferase<br />
yhhM b3467 0.3265 0.0159 putative receptor<br />
yhhN b3468 0.4588 0.0073 putative enzyme<br />
yhhS b3473 0.3667 0.0188 YhhS MFS transporter<br />
yhhT b3474 0.5527 0.0461 hypothetical protein<br />
yhiL b3490 2.2476 0.0051 hypothetical protein<br />
yhiP b3496 1.3365 0.0190 YhiP peptide POT transporter<br />
yhiS b3504 1.6910 0.0222 conserved protein<br />
yhjA b3518 2.6298 0.0076 putative cytochrome C peroxidase (EC 1.11.1)<br />
yhjC b3521 0.2139 0.0092 putative transcriptional regulator LYSR-type<br />
yhjR b3535 0.2767 0.0243 hypothetical protein<br />
yiaJ b3574 0.3464 0.0228 YiaJ transcriptional repressor<br />
yiaV b3586 2.4422 0.0226 putative membrane protein<br />
yibA b3594 2.4261 0.0343 putative lyase<br />
yicE b3654 0.0934 0.0007 YicE NCS2 tranporter<br />
yidI b3677 0.5721 0.0151 hypothetical protein<br />
yieO b3754 0.5670 0.0417 YieO drug MFS transporter<br />
yifE b3764 1.7605 0.0381 hypothetical protein<br />
yigA b3810 0.4061 0.0291 conserved hypothetical protein<br />
yigL b3826 1.9789 0.0325 conserved protein with a phosphatase-like domain<br />
yihI b3866 0.3969 0.0382 conserved protein<br />
yihN b3874 1.3163 0.0362 YihN MFS transporter<br />
yihW b3884 0.5544 0.0009 putative DEOR-type transcriptional regulator<br />
yihX b3885 0.1479 0.0070 putative phosphatase<br />
yjbE b4026 2.1742 0.0103 conserved hypothetical protein<br />
yjbJ b4045 0.8546 0.0153 highly abundant nonessential protein<br />
yjbQ b4056 0.3680 0.0272 conserved hypothetical protein<br />
yjdA b4109 0.1308 0.0055 hypothetical protein<br />
yjeB b4178 0.1691 0.0045 conserved protein with a Winged helix domain<br />
yjeK b4146 0.2195 0.0017 hypothetical protein<br />
yjeS b4166 5.5584 0.0055 conserved ferredoxin-like protein<br />
yjfY b4199 2.3690 0.0232 conserved hypothetical protein<br />
yjfZ b4204 1.7639 0.0428 hypothetical protein<br />
yjgK b4252 0.7295 0.0143 conserved hypothetical protein<br />
135