Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
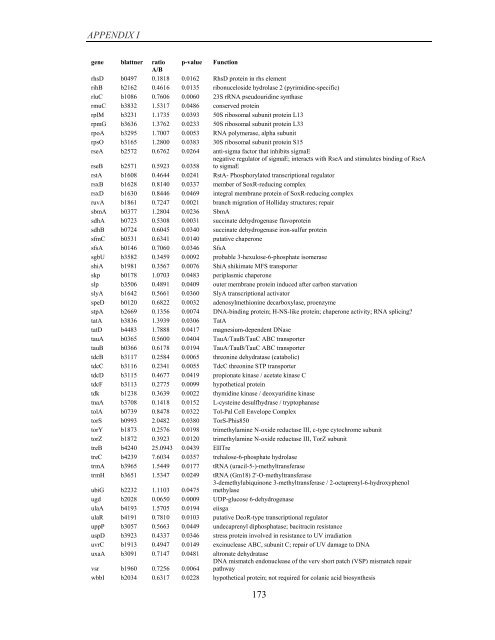
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
rhsD b0497 0.1818 0.0162 RhsD protein in rhs element<br />
rihB b2162 0.4616 0.0135 ribonuceloside hydrolase 2 (pyrimidine-specific)<br />
rluC b1086 0.7606 0.0060 23S rRNA pseudouridine synthase<br />
rmuC b3832 1.5317 0.0486 conserved protein<br />
rplM b3231 1.1735 0.0393 50S ribosomal subunit protein L13<br />
rpmG b3636 1.3762 0.0233 50S ribosomal subunit protein L33<br />
rpoA b3295 1.7007 0.0053 RNA polymerase, alpha subunit<br />
rpsO b3165 1.2800 0.0383 30S ribosomal subunit protein S15<br />
rseA b2572 0.6762 0.0264 anti-sigma factor that inhibits sigmaE<br />
rseB b2571 0.5923 0.0358<br />
negative regulator <strong>of</strong> sigmaE; interacts with RseA and stimulates binding <strong>of</strong> RseA<br />
to sigmaE<br />
rstA b1608 0.4644 0.0241 RstA- Phosphorylated transcriptional regulator<br />
rsxB b1628 0.8140 0.0337 member <strong>of</strong> SoxR-reducing complex<br />
rsxD b1630 0.8446 0.0469 integral membrane protein <strong>of</strong> SoxR-reducing complex<br />
ruvA b1861 0.7247 0.0021 branch migration <strong>of</strong> Holliday structures; repair<br />
sbmA b0377 1.2804 0.0236 SbmA<br />
sdhA b0723 0.5308 0.0031 succinate dehydrogenase flavoprotein<br />
sdhB b0724 0.6045 0.0340 succinate dehydrogenase iron-sulfur protein<br />
sfmC b0531 0.6341 0.0140 putative chaperone<br />
sfsA b0146 0.7060 0.0346 SfsA<br />
sgbU b3582 0.3459 0.0092 probable 3-hexulose-6-phosphate isomerase<br />
shiA b1981 0.3567 0.0076 ShiA shikimate MFS transporter<br />
skp b0178 1.0703 0.0483 periplasmic chaperone<br />
slp b3506 0.4891 0.0409 outer membrane protein induced after carbon starvation<br />
slyA b1642 0.5661 0.0360 SlyA transcriptional activator<br />
speD b0120 0.6822 0.0032 adenosylmethionine decarboxylase, proenzyme<br />
stpA b2669 0.1356 0.0074 DNA-binding protein; H-NS-like protein; chaperone activity; RNA splicing?<br />
tatA b3836 1.3939 0.0306 TatA<br />
tatD b4483 1.7888 0.0417 magnesium-dependent DNase<br />
tauA b0365 0.5600 0.0404 TauA/TauB/TauC ABC transporter<br />
tauB b0366 0.6178 0.0194 TauA/TauB/TauC ABC transporter<br />
tdcB b3117 0.2584 0.0065 threonine dehydratase (catabolic)<br />
tdcC b3116 0.2341 0.0055 TdcC threonine STP transporter<br />
tdcD b3115 0.4677 0.0419 propionate kinase / acetate kinase C<br />
tdcF b3113 0.2775 0.0099 hypothetical protein<br />
tdk b1238 0.3639 0.0022 thymidine kinase / deoxyuridine kinase<br />
tnaA b3708 0.1418 0.0152 L-cysteine desulfhydrase / tryptophanase<br />
tolA b0739 0.8478 0.0322 Tol-Pal Cell Envelope Complex<br />
torS b0993 2.0482 0.0380 TorS-Phis850<br />
torY b1873 0.2576 0.0198 trimethylamine N-oxide reductase III, c-type cytochrome subunit<br />
torZ b1872 0.3923 0.0120 trimethylamine N-oxide reductase III, TorZ subunit<br />
treB b4240 25.0943 0.0439 EIITre<br />
treC b4239 7.6034 0.0357 trehalose-6-phosphate hydrolase<br />
trmA b3965 1.5449 0.0177 tRNA (uracil-5-)-methyltransferase<br />
trmH b3651 1.5347 0.0249 tRNA (Gm18) 2'-O-methyltransferase<br />
ubiG b2232 1.1103 0.0475<br />
3-demethylubiquinone 3-methyltransferase / 2-octaprenyl-6-hydroxyphenol<br />
methylase<br />
ugd b2028 0.0650 0.0009 UDP-glucose 6-dehydrogenase<br />
ulaA b4193 1.5705 0.0194 eiisga<br />
ulaR b4191 0.7810 0.0103 putative DeoR-type transcriptional regulator<br />
uppP b3057 0.5663 0.0449 undecaprenyl diphosphatase; bacitracin resistance<br />
uspD b3923 0.4337 0.0346 stress protein involved in resistance to UV irradiation<br />
uvrC b1913 0.4947 0.0149 excinuclease ABC, subunit C; repair <strong>of</strong> UV damage to DNA<br />
uxaA b3091 0.7147 0.0481 altronate dehydratase<br />
vsr b1960 0.7256 0.0064<br />
DNA mismatch endonuclease <strong>of</strong> the very short patch (VSP) mismatch repair<br />
pathway<br />
wbbI b2034 0.6317 0.0228 hypothetical protein; not required for colanic acid biosynthesis<br />
173