Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
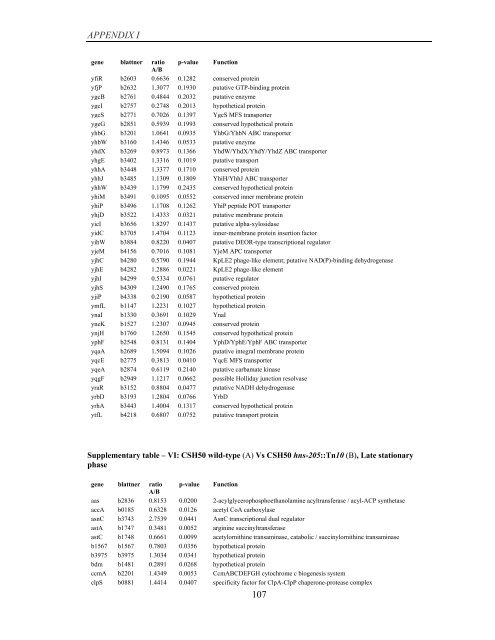
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
yfiR b2603 0.6636 0.1282 conserved protein<br />
yfjP b2632 1.3077 0.1930 putative GTP-binding protein<br />
ygcB b2761 0.4844 0.2032 putative enzyme<br />
ygcI b2757 0.2748 0.2013 hypothetical protein<br />
ygcS b2771 0.7026 0.1397 YgcS MFS transporter<br />
ygeG b2851 0.5939 0.1993 conserved hypothetical protein<br />
yhbG b3201 1.0641 0.0935 YhbG/YhbN ABC transporter<br />
yhbW b3160 1.4346 0.0533 putative enzyme<br />
yhdX b3269 0.8973 0.1366 YhdW/YhdX/YhdY/YhdZ ABC transporter<br />
yhgE b3402 1.3316 0.1019 putative transport<br />
yhhA b3448 1.3377 0.1710 conserved protein<br />
yhhJ b3485 1.1309 0.1809 YhiH/YhhJ ABC transporter<br />
yhhW b3439 1.1799 0.2435 conserved hypothetical protein<br />
yhiM b3491 0.1095 0.0552 conserved inner membrane protein<br />
yhiP b3496 1.1708 0.1262 YhiP peptide POT transporter<br />
yhjD b3522 1.4333 0.0321 putative membrane protein<br />
yicI b3656 1.8297 0.1437 putative alpha-xylosidase<br />
yidC b3705 1.4704 0.1123 inner-membrane protein insertion factor<br />
yihW b3884 0.8220 0.0407 putative DEOR-type transcriptional regulator<br />
yjeM b4156 0.7016 0.1081 YjeM APC transporter<br />
yjhC b4280 0.5790 0.1944 KpLE2 phage-like element; putative NAD(P)-binding dehydrogenase<br />
yjhE b4282 1.2886 0.0221 KpLE2 phage-like element<br />
yjhI b4299 0.5334 0.0761 putative regulator<br />
yjhS b4309 1.2490 0.1765 conserved protein<br />
yjiP b4338 0.2190 0.0587 hypothetical protein<br />
ymfL b1147 1.2231 0.1027 hypothetical protein<br />
ynaI b1330 0.3691 0.1029 YnaI<br />
yneK b1527 1.2307 0.0945 conserved protein<br />
ynjH b1760 1.2650 0.1545 conserved hypothetical protein<br />
yphF b2548 0.8131 0.1404 YphD/YphE/YphF ABC transporter<br />
yqaA b2689 1.5094 0.1026 putative integral membrane protein<br />
yqcE b2775 0.3813 0.0410 YqcE MFS transporter<br />
yqeA b2874 0.6119 0.2140 putative carbamate kinase<br />
yqgF b2949 1.1217 0.0662 possible Holliday junction resolvase<br />
yraR b3152 0.8804 0.0477 putative NADH dehydrogenase<br />
yrbD b3193 1.2804 0.0766 YrbD<br />
yrhA b3443 1.4004 0.1317 conserved hypothetical protein<br />
ytfL b4218 0.6807 0.0752 putative transport protein<br />
Supplementary table – VI: CSH50 wild-type (A) Vs CSH50 hns-205::Tn10 (B), Late stationary<br />
phase<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
aas b2836 0.8153 0.0200 2-acylglycerophosphoethanolamine acyltransferase / acyl-ACP synthetase<br />
accA b0185 0.6328 0.0126 acetyl CoA carboxylase<br />
asnC b3743 2.7539 0.0441 AsnC transcriptional dual regulator<br />
astA b1747 0.3481 0.0052 arginine succinyltransferase<br />
astC b1748 0.6661 0.0099 acetylornithine transaminase, catabolic / succinylornithine transaminase<br />
b1567 b1567 0.7803 0.0356 hypothetical protein<br />
b3975 b3975 1.3034 0.0341 hypothetical protein<br />
bdm b1481 0.2891 0.0268 hypothetical protein<br />
ccmA b2201 1.4349 0.0053 CcmABCDEFGH cytochrome c bio<strong>gene</strong>sis system<br />
clpS b0881 1.4414 0.0407 specificity factor for ClpA-ClpP chaperone-protease complex<br />
107