Smithsonian at the Poles: Contributions to International Polar
Smithsonian at the Poles: Contributions to International Polar
Smithsonian at the Poles: Contributions to International Polar
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
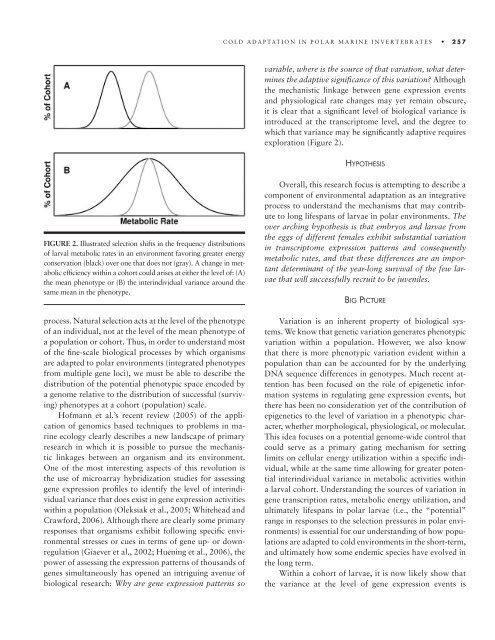
FIGURE 2. Illustr<strong>at</strong>ed selection shifts in <strong>the</strong> frequency distributions<br />
of larval metabolic r<strong>at</strong>es in an environment favoring gre<strong>at</strong>er energy<br />
conserv<strong>at</strong>ion (black) over one th<strong>at</strong> does not (gray). A change in metabolic<br />
effi ciency within a cohort could arises <strong>at</strong> ei<strong>the</strong>r <strong>the</strong> level of: (A)<br />
<strong>the</strong> mean phenotype or (B) <strong>the</strong> interindividual variance around <strong>the</strong><br />
same mean in <strong>the</strong> phenotype.<br />
process. N<strong>at</strong>ural selection acts <strong>at</strong> <strong>the</strong> level of <strong>the</strong> phenotype<br />
of an individual, not <strong>at</strong> <strong>the</strong> level of <strong>the</strong> mean phenotype of<br />
a popul<strong>at</strong>ion or cohort. Thus, in order <strong>to</strong> understand most<br />
of <strong>the</strong> fi ne-scale biological processes by which organisms<br />
are adapted <strong>to</strong> polar environments (integr<strong>at</strong>ed phenotypes<br />
from multiple gene loci), we must be able <strong>to</strong> describe <strong>the</strong><br />
distribution of <strong>the</strong> potential phenotypic space encoded by<br />
a genome rel<strong>at</strong>ive <strong>to</strong> <strong>the</strong> distribution of successful (surviving)<br />
phenotypes <strong>at</strong> a cohort (popul<strong>at</strong>ion) scale.<br />
Hofmann et al.’s recent review (2005) of <strong>the</strong> applic<strong>at</strong>ion<br />
of genomics based techniques <strong>to</strong> problems in marine<br />
ecology clearly describes a new landscape of primary<br />
research in which it is possible <strong>to</strong> pursue <strong>the</strong> mechanistic<br />
linkages between an organism and its environment.<br />
One of <strong>the</strong> most interesting aspects of this revolution is<br />
<strong>the</strong> use of microarray hybridiz<strong>at</strong>ion studies for assessing<br />
gene expression profi les <strong>to</strong> identify <strong>the</strong> level of interindividual<br />
variance th<strong>at</strong> does exist in gene expression activities<br />
within a popul<strong>at</strong>ion (Oleksiak et al., 2005; Whitehead and<br />
Crawford, 2006). Although <strong>the</strong>re are clearly some primary<br />
responses th<strong>at</strong> organisms exhibit following specifi c environmental<br />
stresses or cues in terms of gene up- or downregul<strong>at</strong>ion<br />
(Giaever et al., 2002; Huening et al., 2006), <strong>the</strong><br />
power of assessing <strong>the</strong> expression p<strong>at</strong>terns of thousands of<br />
genes simultaneously has opened an intriguing avenue of<br />
biological research: Why are gene expression p<strong>at</strong>terns so<br />
COLD ADAPTATION IN POLAR MARINE INVERTEBRATES 257<br />
variable, where is <strong>the</strong> source of th<strong>at</strong> vari<strong>at</strong>ion, wh<strong>at</strong> determines<br />
<strong>the</strong> adaptive signifi cance of this vari<strong>at</strong>ion? Although<br />
<strong>the</strong> mechanistic linkage between gene expression events<br />
and physiological r<strong>at</strong>e changes may yet remain obscure,<br />
it is clear th<strong>at</strong> a signifi cant level of biological variance is<br />
introduced <strong>at</strong> <strong>the</strong> transcrip<strong>to</strong>me level, and <strong>the</strong> degree <strong>to</strong><br />
which th<strong>at</strong> variance may be signifi cantly adaptive requires<br />
explor<strong>at</strong>ion (Figure 2).<br />
HYPOTHESIS<br />
Overall, this research focus is <strong>at</strong>tempting <strong>to</strong> describe a<br />
component of environmental adapt<strong>at</strong>ion as an integr<strong>at</strong>ive<br />
process <strong>to</strong> understand <strong>the</strong> mechanisms th<strong>at</strong> may contribute<br />
<strong>to</strong> long lifespans of larvae in polar environments. The<br />
over arching hypo<strong>the</strong>sis is th<strong>at</strong> embryos and larvae from<br />
<strong>the</strong> eggs of different females exhibit substantial vari<strong>at</strong>ion<br />
in transcrip<strong>to</strong>me expression p<strong>at</strong>terns and consequently<br />
metabolic r<strong>at</strong>es, and th<strong>at</strong> <strong>the</strong>se differences are an important<br />
determinant of <strong>the</strong> year-long survival of <strong>the</strong> few larvae<br />
th<strong>at</strong> will successfully recruit <strong>to</strong> be juveniles.<br />
BIG PICTURE<br />
Vari<strong>at</strong>ion is an inherent property of biological systems.<br />
We know th<strong>at</strong> genetic vari<strong>at</strong>ion gener<strong>at</strong>es phenotypic<br />
vari<strong>at</strong>ion within a popul<strong>at</strong>ion. However, we also know<br />
th<strong>at</strong> <strong>the</strong>re is more phenotypic vari<strong>at</strong>ion evident within a<br />
popul<strong>at</strong>ion than can be accounted for by <strong>the</strong> underlying<br />
DNA sequence differences in genotypes. Much recent <strong>at</strong>tention<br />
has been focused on <strong>the</strong> role of epigenetic inform<strong>at</strong>ion<br />
systems in regul<strong>at</strong>ing gene expression events, but<br />
<strong>the</strong>re has been no consider<strong>at</strong>ion yet of <strong>the</strong> contribution of<br />
epigenetics <strong>to</strong> <strong>the</strong> level of vari<strong>at</strong>ion in a phenotypic character,<br />
whe<strong>the</strong>r morphological, physiological, or molecular.<br />
This idea focuses on a potential genome-wide control th<strong>at</strong><br />
could serve as a primary g<strong>at</strong>ing mechanism for setting<br />
limits on cellular energy utiliz<strong>at</strong>ion within a specifi c individual,<br />
while <strong>at</strong> <strong>the</strong> same time allowing for gre<strong>at</strong>er potential<br />
interindividual variance in metabolic activities within<br />
a larval cohort. Understanding <strong>the</strong> sources of vari<strong>at</strong>ion in<br />
gene transcription r<strong>at</strong>es, metabolic energy utiliz<strong>at</strong>ion, and<br />
ultim<strong>at</strong>ely lifespans in polar larvae (i.e., <strong>the</strong> “potential”<br />
range in responses <strong>to</strong> <strong>the</strong> selection pressures in polar environments)<br />
is essential for our understanding of how popul<strong>at</strong>ions<br />
are adapted <strong>to</strong> cold environments in <strong>the</strong> short-term,<br />
and ultim<strong>at</strong>ely how some endemic species have evolved in<br />
<strong>the</strong> long term.<br />
Within a cohort of larvae, it is now likely show th<strong>at</strong><br />
<strong>the</strong> variance <strong>at</strong> <strong>the</strong> level of gene expression events is