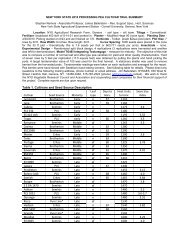
conference schedule and program with abstracts - Horticulture ...
conference schedule and program with abstracts - Horticulture ...
conference schedule and program with abstracts - Horticulture ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
P-33<br />
Analysis of expressed sequence tags (EST) from Vitis vinifera L. flower <strong>and</strong> fruit <strong>and</strong><br />
electronic mapping of new ESTs<br />
J. Fang*, L. Shangguan, C. Song, C. Wang, N.K. Korir, G. Yang<br />
College of <strong>Horticulture</strong>, Nanjing Agricultural University, Nanjing, China<br />
*Presenting author: fanggg@njau.edu.cn<br />
Expressed Sequence Tags (EST) are a cost-effective molecular biology tool that eases our<br />
underst<strong>and</strong>ing of genes involved in plant growth <strong>and</strong> differentiation. ESTs represent a valuable<br />
<strong>and</strong> abundant resource for genome annotation, gene expression, <strong>and</strong> comparative genomics in<br />
plants. In this study, a total of 6,230 EST sequences were produced from 7,561 clones in the<br />
grape (Vitis vinifera cv. Summer Black) flower <strong>and</strong> fruit cDNA library. After cluster <strong>and</strong><br />
assembly of datasets, 3,582 unigenes comprising of 1,070 contigs <strong>and</strong> 2,512 singlets were<br />
established. 925 unique ESTs <strong>with</strong> known or putative functions were gained by BlastX analysis<br />
against the NCBI non-redundant protein database (nr). The sequences were assigned to 12<br />
putative cellular roles based on additional functional annotation as well as comparative <strong>and</strong><br />
classification analysis. 381 new ESTs were gained after Blastn analysis, <strong>and</strong> 79 SSR loci were<br />
mined from these new ESTs by MISA. After electronic mapping, 289 new EST sequences could<br />
be mapped to all the 19 grape chromosomes. The three highest matching chromosomes (chr)<br />
were chr18, chr5 <strong>and</strong> chr14, <strong>with</strong> 11.76% (34), 8.30% (24) <strong>and</strong> 7.96% (23) new ESTs mapped<br />
on, respectively.<br />
109