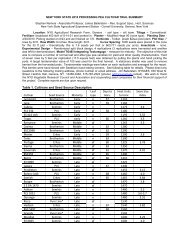
conference schedule and program with abstracts - Horticulture ...
conference schedule and program with abstracts - Horticulture ...
conference schedule and program with abstracts - Horticulture ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
P-61<br />
Grepevine variety determination from herbarium <strong>and</strong> arheological specimens<br />
N. Malenica 1 , S. Simon 2 , E. Maletic 3 , I. Pejic* 2<br />
1<br />
University of Zagreb, Faculty of Science, Department of Molecular Biology, Zagreb, Croatia; 2<br />
University of Zagreb, Faculty of Agriculture, Department of Plant Breeding, Genetics <strong>and</strong><br />
Biometrics, Zagreb, Croatia; 3 University of Zagreb, Faculty of Agriculture, Department of<br />
Viticulture <strong>and</strong> Enology, Zagreb, Croatia<br />
*Corresponding author: ipejic@agr.hr<br />
Genotyping old grapevine samples, in particular variety identification via microsatellite profiling<br />
is still far from being routine. The successful identification depends on several factors, the age of<br />
the investigated material being the most obvious one. In addition, the amount <strong>and</strong> integrity of<br />
DNA depends on conditions of samples storing. Contamination <strong>and</strong> fragmentation processes<br />
make the isolated ancient DNA (aDNA) a rather difficult template for PCR amplification. Three<br />
old grapevine samples were investigated in this study. Two samples were taken from<br />
approximately 90-years old herbarium of cv. Tribidrag, a Croatian autochthonous grapevine<br />
variety not anymore present in the germplasm collections <strong>and</strong> one was approximately 2000 years<br />
old underwater archeological grapevine specimen (woody parts). Several different approaches of<br />
DNA isolation were tested. However, the use of a commercial plant DNA isolation kit showed to<br />
be the best choice in terms of yield <strong>and</strong> prevention of possible contamination <strong>with</strong> temporary<br />
grapevine DNA. The initial attempts to amplify st<strong>and</strong>ard grapevine microsatellite loci by<br />
st<strong>and</strong>ard PCR protocol failed, most likely due to low copy number of template DNA. Therefore,<br />
the whole genome amplification (WGA) kit was successfully applied to overcome this problem.<br />
The WGA approach worked in case of one herbarium sample <strong>and</strong> the archeological specimen.<br />
DNA fragments ranging between 100 <strong>and</strong> 500 bp were cloned <strong>and</strong> sequenced. While the isolated<br />
DNA from the underwater archeological sample was proved to be from different origins (not<br />
from grapevine), indicating contamination of the specimen, the six sequenced clones from the<br />
herbarium specimen corresponded to six different grapevine chromosomes. Therefore, we used<br />
the WGA-processed herbarium sample <strong>and</strong> managed to PCR-amplify the VVS2 locus, thereby<br />
demonstrating the proof of principle. Eventually, we successfully genotyped the same template<br />
using six st<strong>and</strong>ard microsatellite loci (VVS2, VVMD5, VVMD7, VVMD27, VvZAG62 <strong>and</strong><br />
VvZAG79). The obtained SSR profile was identical to Zinf<strong>and</strong>el/Primitivo/Crljenak Kastelanski,<br />
except for the VVS2 locus which was homozygous (141/141) instead of heterozygous (131/141).<br />
In the light of these results we hypothesize on the origin of Zinf<strong>and</strong>el as an old Croatian variety<br />
under the historical name Tribidrag, which was being used in documents dating as late as 15 th<br />
century.<br />
140