THESE Maryse Bonnin Jusserand - Université de Bourgogne
THESE Maryse Bonnin Jusserand - Université de Bourgogne
THESE Maryse Bonnin Jusserand - Université de Bourgogne
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
1/Vi<br />
25<br />
20<br />
15<br />
10<br />
5<br />
0<br />
y = 1,820x + 1,773<br />
R² = 0,999<br />
0 2 4 6 8 10 12<br />
Résultats<br />
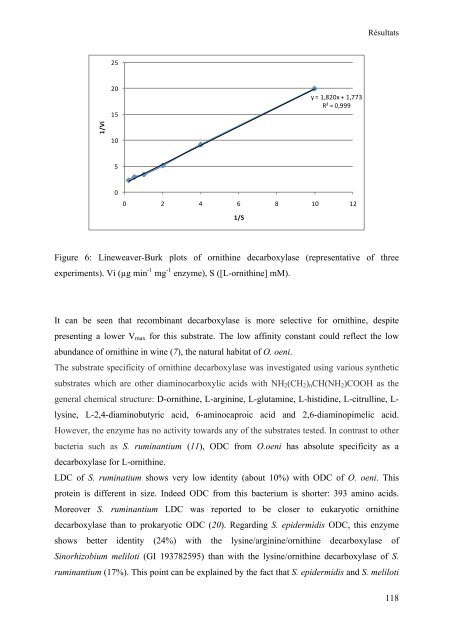
Figure 6: Lineweaver-Burk plots of ornithine <strong>de</strong>carboxylase (representative of three<br />
experiments). Vi (µg min -1 mg -1 enzyme), S ([L-ornithine] mM).<br />
It can be seen that recombinant <strong>de</strong>carboxylase is more selective for ornithine, <strong>de</strong>spite<br />
presenting a lower Vmax for this substrate. The low affinity constant could reflect the low<br />
abundance of ornithine in wine (7), the natural habitat of O. oeni.<br />
The substrate specificity of ornithine <strong>de</strong>carboxylase was investigated using various synthetic<br />
substrates which are other diaminocarboxylic acids with NH2(CH2)nCH(NH2)COOH as the<br />
general chemical structure: D-ornithine, L-arginine, L-glutamine, L-histidine, L-citrulline, L-<br />
lysine, L-2,4-diaminobutyric acid, 6-aminocaproic acid and 2,6-diaminopimelic acid.<br />
However, the enzyme has no activity towards any of the substrates tested. In contrast to other<br />
bacteria such as S. ruminantium (11), ODC from O.oeni has absolute specificity as a<br />
<strong>de</strong>carboxylase for L-ornithine.<br />
LDC of S. ruminatium shows very low i<strong>de</strong>ntity (about 10%) with ODC of O. oeni. This<br />
protein is different in size. In<strong>de</strong>ed ODC from this bacterium is shorter: 393 amino acids.<br />
Moreover S. ruminantium LDC was reported to be closer to eukaryotic ornithine<br />
<strong>de</strong>carboxylase than to prokaryotic ODC (20). Regarding S. epi<strong>de</strong>rmidis ODC, this enzyme<br />
shows better i<strong>de</strong>ntity (24%) with the lysine/arginine/ornithine <strong>de</strong>carboxylase of<br />
Sinorhizobium meliloti (GI 193782595) than with the lysine/ornithine <strong>de</strong>carboxylase of S.<br />
ruminantium (17%). This point can be explained by the fact that S. epi<strong>de</strong>rmidis and S. meliloti<br />
1/S<br />
118